Hi,
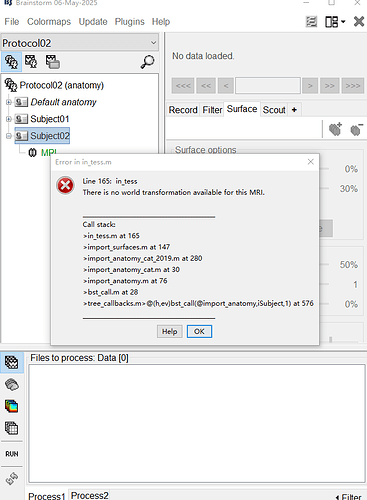
I am using the latest Brainstorm in MATLAB 2023. T1 MRI can be segmented by Cat12 (also the latest). But it fails to import the anatomy from Cat12 folder into Brainstorm. The error is "There is no world transformation available for this MRI". Anyone knows what's the problem?
Thanks
- Can you confirm the version and release of CAT12 that you are using?
[CATrel, CATver] = cat_version
The fact that the anatomy folder is imported with import_anatomy_cat_2019 (instead of 2020) indicates that you may be running an older version of CAT12.
Also, it seems your NIfTI file is really missing the initial transformation from voxels to world.
This is stored in the sform and qform fields.
Can you check if this transformation is indeed not in the MRI file?
Open the MRI file with a MRI software, e.g. FSL
Thank you so much for the reply.
1.The answer of runing [CATrel, CATver] = cat_version is
CATrel ='CAT12.9'
CATver = '2577'
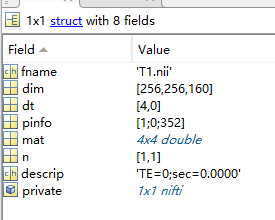
- I open the T1.nift file with "vol = spm_vol('T1.nii');“,
3.Yes, CAT12 can be called in Brainstorm and segmentation is successful.
Thanks
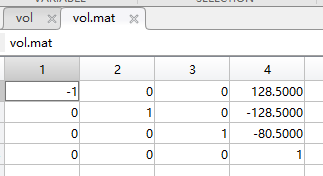
The data in the field of mat:
Does this mean the trouble you are having is when importing a CAT12 folder that was not segmented from Brainstorm?
Yes, that's what I meant. Previously, I used CAT12 run segmentation first. Then I tried to import the result folder into Brainstorm, but it failed.
Raymundo,
I used another T1.nift with sform and qform for segmentaiton, and the result folder can be imported into Brainstorm successfully.
Thank you so much for your help.