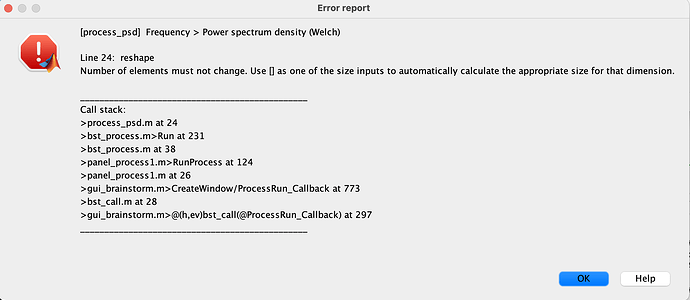
Dear Brainstorm users, I am getting this error message in two subject files from my MEG data set (recorded on Elekta, Neuromag System).
I have tried to re-transfer these files a few times, I am still getting this message. Any help here would be useful, thanks in advance ![]()
It looks like the problem comes from the computation of the PSD, not the import of the FIF files.
- Can you open the MEG recordings for display? Please include a screen capture here.
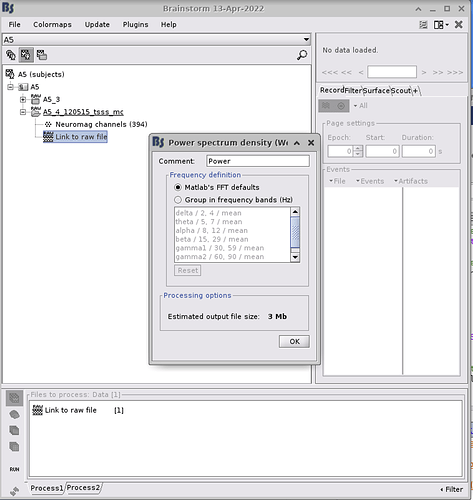
- Please post a screen capture of the PSD options you are using
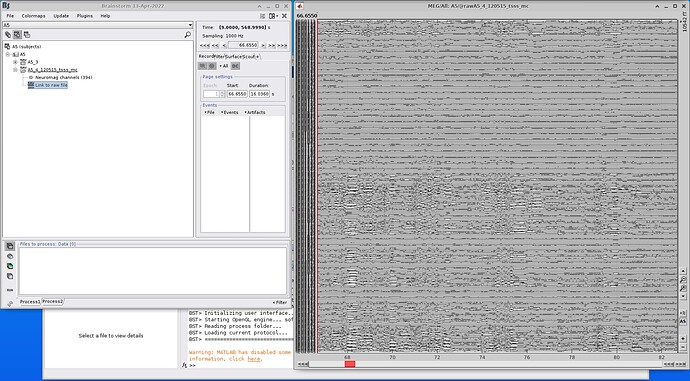
Yes, you are correct! I am able to import the MEG file individually - screenshot attached (I had created a process for importing and computing PSD which was failing, therefore the confusion).
I am getting an error when trying to compute the PSD. screenshots attached below:
What you show should be working.
Further debugging:
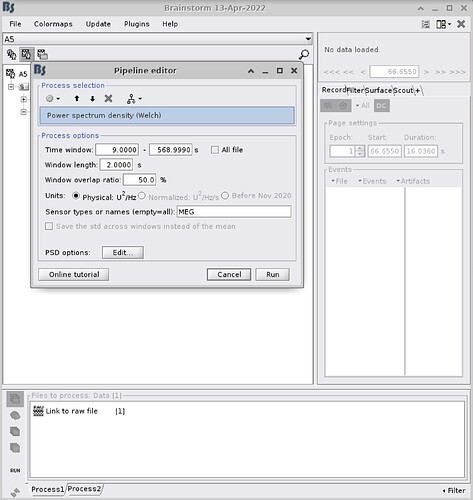
- Can you post a screen capture of the PSD frequency options (click on the Edit button in the options panel)? Make sure the "Matlab's FFT defaults" option is selected.
- Make sure you don't have any other Matlab toolboxes interfering: Close Brainstorm, edit your Matlab path, remove all the folders that are not related to the Matlab installation itself. Restart Matlab and Brainstorm, try again.
If you can't get this to work, can you please share an example data file?
- Right-click on subject A5 > File > Duplicate
- In the copy: Right-click on the tsss Link to raw file > Import in database > Import 5 second of recordings, with no events, no baseline correction, no new folder, no resampling.
- In the copy: Delete the RAW folders, keep only the 5s imported block with its channel file.
- Run the PSD computation on this 5s block. Make sure it produces exactly the same error as the continuous files (the error message you reported).
- Right-click on the subject copy folder > File > Export subject
- Upload the .zip file somewhere and post the download link here.
Thanks a lot for your reply Francois! Here is a screenshot of the PSD options.
I had tried your second suggestion several times before posting the question, and I had also tried running this file in another MATLAB from a completely different system, and the same error popped up.
I tried importing the data and snipping off the last few seconds of the recording, and the PSD and filtering worked! But now for some reason, the SSP is disabled for this subject!
Here is the link to the subject files A5_new_A5.zip - Google Drive
I am facing this issue with only 2 subjects in my study cohort, but since my sample size is limited, every subject is critical. So any suggestion to resolve this would be very helpful!!
thanks in advance ![]()
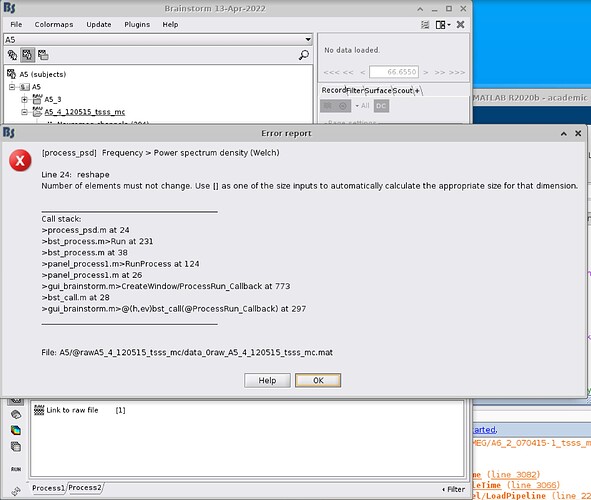
Unfortunately, I can't reproduce the error with the Brainstorm protocol you shared...
It does not include the original FIF file (only the link to it), and if I use the continuous blocks of data you imported to the database: the PSD computation works.
If you'd like me to try to debug this issue: please share the original .fif file instead of a Brainstorm protocol.
Please make sure that all the information you share relate to the same dataset. Here, it does not seem to be the case. You shared a file defined for [1492s - 2061s], while your first screen captures showed different time windows (9s - 569s).
I tried importing the data and snipping off the last few seconds of the recording, and the PSD and filtering worked! But now for some reason, the SSP is disabled for this subject!
All the dynamic linear operations (SSP, ICA, re-referencing) are only available on continuous files.
Avoid import large segments of recordings to the database, most of the Brainstorm display and computation functions are not meant to work like this. First do all you need to do on the continuous files (cleaning, filtering...) and then epoch your data.
https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Review_vs_Import
Hi, please find attached the two .fif files from the two subjects which are having this particular issue. Apologies for the confusion on data sets. The [1429, 2061] and [9, 569] are two runs of the same subject- both were showing this error.
Point noted on importing files to the database.
Thank you for the example files.
I tested only the first one (A5), but I guess the problem is the same for both.
There is something wrong with these FIF files, which has nothing to do with Brainstorm. There are some missing samples at the end of the last data block. If you double-click on the file to review the signals and then navigate to the very end of the file, you would generate an error as well. As you noticed, you can get rid of the error by selected a shorter time window in the PSD.
I added a test for this specific case, so that instead of crashing, the reading function returns a data matrix padded with zeros: https://github.com/brainstorm-tools/brainstorm3/commit/695b4379e6fbfd493d8294b38cf44e4e20615903
It displays a message in the Matlab command window, when trying to read the end of the file:
FIF> Error: Missing samples #2061.997000s to #2061.999000s. Padding with zeros...
Update Brainstorm to get this fix.
However, you should not rely on this fix for your analysis: the missing data is still missing, even though now it is going to be silent: You still need to exclude the last few milliseconds from the PSD computation in order to minimize the effect on your spectrum estimation of these added zeros.
You might find other processes that need to be fixed to work correctly with these files.
If you find any, please report them here as well.