Hello,
I was trying to export some preprocessed HCP MEG data to MATLAB in .mat format. When I tried to do this, it gave me an error saying that I could not do this, but that I could request this feature in the forums here.
If exporting this data as a .mat file is not possible for now, I have a follow up question. I can export the data as a .bst file, but I then want to use it in the 'in_bst_results' which currently throws an error at the .bst file saying that my input must be a MAT file or an ASCII file, so are there any good ways to convert a .bst to a .mat or .ascii?
I appreciate the help!
-Nikolai
Raw MEG data in the Human Connectome Project (HCP) ConnectomeDB database is saved as 4D Neuroimaging which is a format that is supported in Brainstorm.
Check this tutorial based on the HCP data.
https://neuroimage.usc.edu/brainstorm/Tutorials/HCP-MEG
 The operations in that tutorial assume familiarity with Brainstorm. If you are a new Brainstorm user, we recommend you start by following the introduction tutorials first (section "Get started" on the tutorial page), using the example dataset that is provided.
The operations in that tutorial assume familiarity with Brainstorm. If you are a new Brainstorm user, we recommend you start by following the introduction tutorials first (section "Get started" on the tutorial page), using the example dataset that is provided.
https://neuroimage.usc.edu/brainstorm/Tutorials
About exporting, once data is in Brainstorm it is possible to export it in other formats.
https://neuroimage.usc.edu/brainstorm/Tutorials/Scripting#Export.2FImport_with_the_database_explorer
Thank you for the reply, Raymundo. I am working with Nikolai on this dataset.
The big picture is that we are trying to save the computed full source estimation results. We have been following the HCP tutorial, but it ends before demonstrating the save full results step. We have also been following the base MEG source estimation tutorial and other scripting tutorials, but unfortunately they have sent us down a few winding paths that we have not been able to sort out. We hope you can help us troubleshoot this by providing detailed steps to resolve this issue that we have not been able to find in the tutorials.
These are the steps we followed:
-
From the raw data file, we selected "Import in database". From there we clicked "Import". If we read correctly, this was recommended by the Source Estimation tutorial, because computing full results doesn't work on raw files. We used the default parameters in the next window because we want a single epoch of data.
-
In the new imported file, we selected "Compute head model" and proceeded with the default settings for Overlapping spheres.
-
Next, we selected "Compute sources [2018]" from the right click menu. Then we selected "Full results (Kernel*Recordings".
-
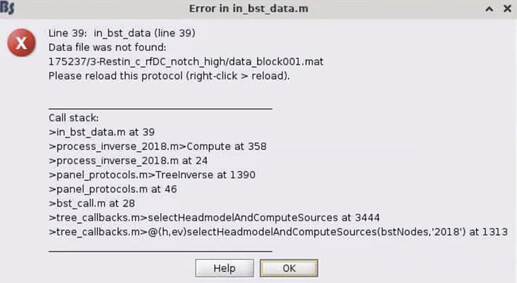
Then, we get an error in in_bst_data (see below). There is a data_block001.mat that could not be found and was never created in our 3-Restin... folder, but the tutorials are not clear about which step creates this file. However, it does tell us to reload this protocol. (This error happens also if we select the inverse kernel option on the last window.)
-
We followed this instruction to reload the protocol. After doing so, we noticed that the raw recording under the imported data file vanishes.
-
We tried "Compute sources [2018]" again. Here, we notice that the "Full results" bubble is grayed out. Compute the inverse kernel works this time, however.
In conclusion, the in_bst_data error caused by the missing data_block001.mat seems to be sending us into a path of failure. Did we make an erroneous step previously?
If there is a better venue (i.e. office hours) to discuss this problem with you, please don't hesitate to reach out to us.
Thanks again,
David