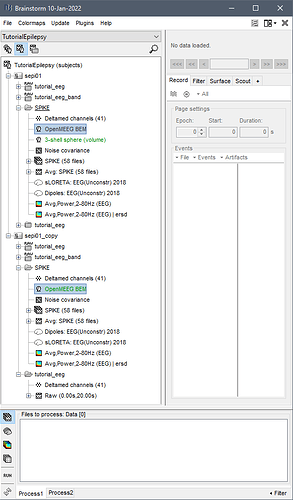
I'm trying to export my traceability results based on the roi I made, but this error window pops up and I need to click confirm manually.
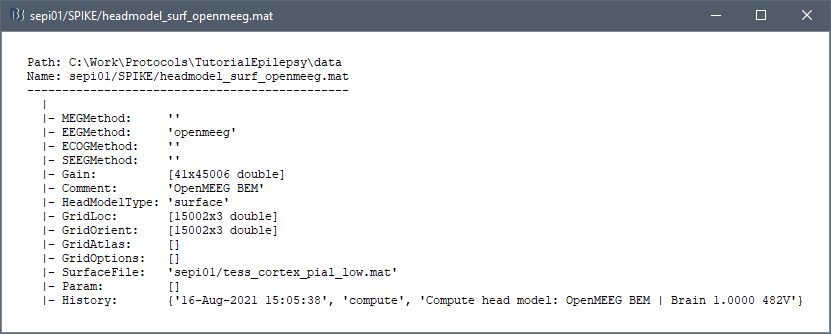
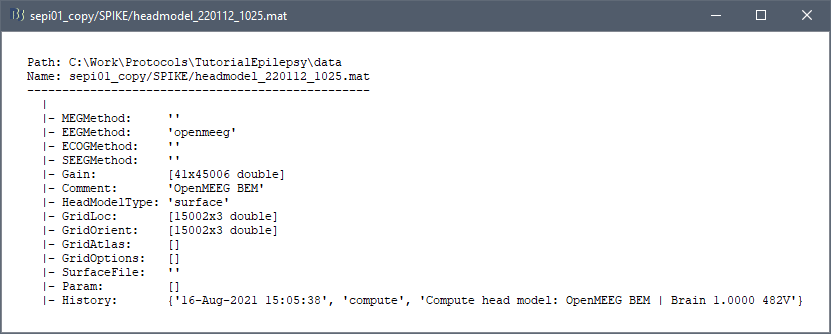
The cortical file associated with these sources no longer exists. Please use: s001_tess_cortex.mat.
I would like to ask if there is any solution for this?
Q1: I first want to ask if it is possible to generate source results with cortex files after preprocessing, or roi exported data with cortex files?
Because I have more than 100 subjects, each subject has 12 evoke files preprocessed in MNE and imported into brainstorm.
My subjects are eeg data of different children from 7-18 years old. So I calculated the brain templates and open meeg bem head models for children aged 7-18 years in advance, and then copied the brain templates and bem models for each subject of the corresponding age. If the bem is regenerated for each subject, there is no such problem. But I have more than 100 subjects, and it takes too long to regenerate bem for each subject.
2 Is there a code to fix this popup or blank cortex file? The one in the previous forum doesn't seem to work.
Thank you very much for your help!
Looking forward to your reply!
Appendix 1:
I have found people with the same problem in the forum
But it keeps reporting errors when following this code to make changes.
[sMat, matName] = in_bst(sFiles{i});
ProtocolInfo = bst_get('ProtocolInfo');
sMat.SurfaceFile = 'SubjectName/cortexfile.mat'; % modify this line
outputFile = bst_fullfile(ProtocolInfo.STUDIES, sFiles{i})
bst_save(outputFile, sMat, 'v6').
It seems that the person who asked this question didn't succeed in using this code, and ended up using AutoHotKey to solve the problem. But I'm using ubuntu and can't use this solution.
This is my code.
Subj={
's001',
};
for i=1:length(Subj)
% Input files
sFiles = [];
SubjectNames = {Subj{i}};
RawFiles = {...
strcat( '/home/jade/data/loopvis/nh_evoke/',Subj{i}, 'evokev1-epo.fif'), ...
strcat( '/home/jade/data/loopvis/nh_evoke/',Subj{i}, 'evokev2-epo.fif'), ...
strcat( '/home/jade/data/loopvis/nh_evoke/',Subj{i}, 'evokev3-epo.fif'), ...
strcat( '/home/jade/data/loopvis/nh_evoke/',Subj{i}, 'evokev4-epo.fif'), };
% Start a new report
bst_report('Start', sFiles);
% Processing. Import MEG/EEG: existing ephemeral data
sFiles = bst_process('CallProcess', 'process_import_data_epoch', sFiles, [], ...
'subjectname', SubjectNames{1}, ...
'condition', '', ...
'dataFiles', {{RawFiles{1}; RawFiles{2}; RawFiles{3}; RawFiles{4}}, 'FIF'}, ...
'iepochs', [], ...
'eventtypes', '', ...
'createcond', 0, ...
'channelalign', 0, ...
'usectfcomp', 0, ...
'usessp', 0, ...
'freq', [], ...
'baseline', []).
[sMat, matName] = in_bst(sFiles);
ProtocolInfo = bst_get('ProtocolInfo');
sMat.SurfaceFile = 'n001/cortexfile.mat'; % change this line
outputFile = bst_fullfile(ProtocolInfo.STUDIES, sFiles{i})
bst_save(outputFile, sMat, 'v6').
% Save and display the report
ReportFile = bst_report('Save', sFiles);
bst_report('Open', ReportFile);
% bst_report('Export', ReportFile, ExportDir);
End
This one is reporting an error
[sMat, matName] = in_bst(sFiles);
The output parameter "sMatrix" (and possibly other parameters) is not assigned when calling "in_bst".
outputFile = bst_fullfile(ProtocolInfo.STUDIES, sFiles{i})
Brace indexing is not supported for this type of variable.
Appendix 2:
This is all my processing scripts
Subj={ 's001',
};
for i=1:length(Subj)
% Input files
sFiles = [];
SubjectNames = {Subj{i}};
RawFiles = {...
strcat( '/home/jade/data/loopvis/ci_evoke/',Subj{i},'evokev1-epo.fif'), ...
strcat( '/home/jade/data/loopvis/ci_evoke/',Subj{i},'evokev2-epo.fif'), ...
strcat( '/home/jade/data/loopvis/ci_evoke/',Subj{i},'evokev3-epo.fif'), ...
strcat( '/home/jade/data/loopvis/ci_evoke/',Subj{i},'evokev4-epo.fif'), };
% Start a new report
bst_report('Start', sFiles);
% Process: Import MEG/EEG: Existing epochs
sFiles = bst_process('CallProcess', 'process_import_data_epoch', sFiles, [], ...
'subjectname', SubjectNames{1}, ...
'condition', '', ...
'datafile', {{RawFiles{1}; RawFiles{2}; RawFiles{3}; RawFiles{4}}, 'FIF'}, ...
'iepochs', [], ...
'eventtypes', '', ...
'createcond', 0, ...
'channelalign', 0, ...
'usectfcomp', 0, ...
'usessp', 0, ...
'freq', [], ...
'baseline', []);
% Process: Set channel file
sFiles = bst_process('CallProcess', 'process_import_channel', sFiles, [], ...
'channelfile', {RawFiles{1}; RawFiles{2}; RawFiles{3}; RawFiles{4}}, ...
'usedefault', 114, ... % NotAligned: GSN HydroCel 128 E1
'channelalign', 1, ...
'fixunits', 1, ...
'vox2ras', 1);
% Process: Set channels type
sFiles = bst_process('CallProcess', 'process_channel_settype', sFiles, [], ...
'sensortypes', 'E8,E14,E21,E25,E125,E126,E127,E128', ...
'newtype', 'EOG');
% Process: Average: By trial group (folder average)
sFiles = bst_process('CallProcess', 'process_average', sFiles, [], ...
'avgtype', 5, ... % By trial group (folder average)
'avg_func', 1, ... % Arithmetic average: mean(x)
'weighted', 0, ...
'keepevents', 0);
% Process: Compute covariance (noise or data)
sFiles = bst_process('CallProcess', 'process_noisecov', sFiles, [], ...
'baseline', [-0.1, 0], ...
'datatimewindow', [0, 0.5], ...
'sensortypes', 'EEG', ...
'target', 1, ... % Noise covariance (covariance over baseline time window)
'dcoffset', 1, ... % Block by block, to avoid effects of slow shifts in data
'identity', 0, ...
'copycond', 0, ...
'copysubj', 0, ...
'copymatch', 0, ...
'replacefile', 3, ... % Keep
'source_abs', 0);
% Process: Compute sources [2018]
sFiles = bst_process('CallProcess', 'process_inverse_2018', sFiles, [], ...
'output', 2, ... % Kernel only: one per file
'inverse', struct(...
'Comment', 'sLORETA: EEG', ...
'InverseMethod', 'minnorm', ...
'InverseMeasure', 'sloreta', ...
'SourceOrient', {{'fixed'}}, ...
'Loose', 0.2, ...
'UseDepth', 0, ...
'WeightExp', 0.5, ...
'WeightLimit', 10, ...
'NoiseMethod', 'reg', ...
'NoiseReg', 0.1, ...
'SnrMethod', 'fixed', ...
'SnrRms', 1e-06, ...
'SnrFixed', 3, ...
'ComputeKernel', 1, ...
'DataTypes', {{'EEG'}}));
% Save and display report
ReportFile = bst_report('Save', sFiles);
bst_report('Open', ReportFile);
% bst_report('Export', ReportFile, ExportDir);
end
% Script generated by Brainstorm (28-Dec-2021)
% Input files
sFiles = {...
'link|s001/s001evokev1-epo/results_sLORETA_EEG_KERNEL_220105_0928.mat|s001/s001evokev1-epo/data_Epoch_average_220105_0916.mat'};
% Start a new report
bst_report('Start', sFiles);
% Process: Scouts time series: Cortex L Cortex R
sFiles = bst_process('CallProcess', 'process_extract_scout', sFiles, [], ...
'timewindow', [-0.1, 0.5], ...
'scouts', {'Structures', {'Cortex L', 'Cortex R'}}, ...
'scoutfunc', 1, ... % Mean
'isflip', 1, ...
'isnorm', 0, ...
'concatenate', 1, ...
'save', 1, ...
'addrowcomment', 1, ...
'addfilecomment', 1);
% Save and display report
ReportFile = bst_report('Save', sFiles);
bst_report('Open', ReportFile);