Hi!
I have been trying to export the raw eeg files to edf format in BS, but I can't make it,
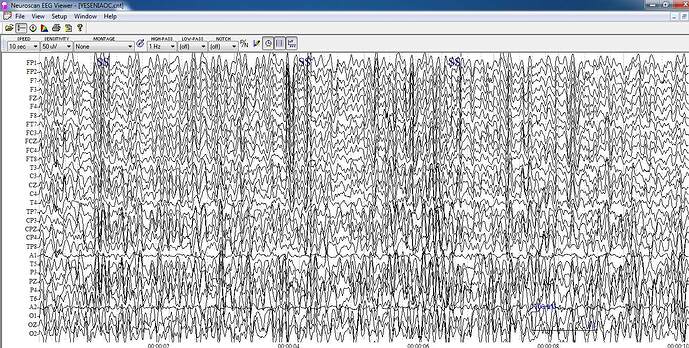
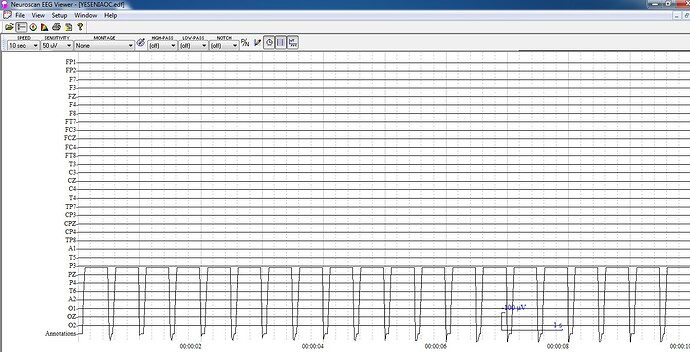
I just select the "review eeg raw file" option in order to import my file, then, when I choose to export it in edf file it seems to work but the files are so diferent, the edf exported file seems to be flat, I attach here the original file and the edf exported file.
Thanks in advance!
pd: do someone has been able to export raw eeg to edf succesfully?
Original:
BS Exported:
The channel "Annotations" contains the events information. This is not EEG data and should not be displayed together with the EEG channels, because its values are much higher it makes all the other signals appear as flat.
It looks like the Neuroscan browser does not interpret it correctly. Maybe it supports the old EDF format, but not the full EDF+ specification...
One solution would be for you to simply hide the Annotations channel in the Neuroscan viewer.
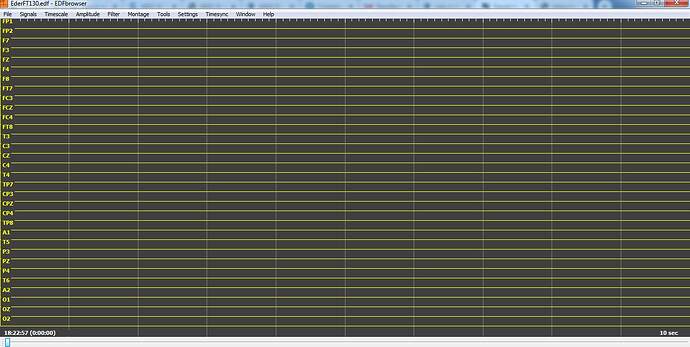
If you want to double-check that the conversion is correct, install EDFbrowser:
https://www.teuniz.net/edfbrowser/
Why are you not exporting to EDF directly using the Neuroscan software?
Thanks for the reply!
Well, that is the thing: the original EEG data does not contain any "Annotations" channel, it was added with the edf exportation process, and in the images, both files have the same scale (50 µV), so even if I hide the annotation channel signal stills flat.
I tryed with another EEG file and I check it on EDF Browser, this time there was no annotation channel, but signal is flat again, no mather if I change zoom or scale

Can you share a short example .cnt file ?
(upload it somewhere and post the download link here)
Why are you not doing the EDF export from the Neuroscan software directly?
Here is the file:
I tryied to use another file (.bin) wich is in the tutorial of epilepsy and it exported the edf file correctly, so the problem seems to be neuroscan format.
Yes, I think it is a better idea to use Neuroscan software to export to edf, I'm just courious about if once the eeg raw file is clean and modified (filtered, maybe a channel renamed of deleted) if I could get the data back in to a continous file and use it with neuroscan (or another EDF viewers) that could be so useful, specially in clinical sets.
But, yes, it is not a real problem.
The export of this file to EDF works well with this file.
For some reason, the automatic scaling of EDFbrowser is weird. Select the menu Amplitude>Fit to pane, and you will see your signals.
Otherwise, link the exported EDF file again into Brainstorm, and import 1s of each (the CNT and the EDF files) and compare them (left=original CNT, right=exported EDF):
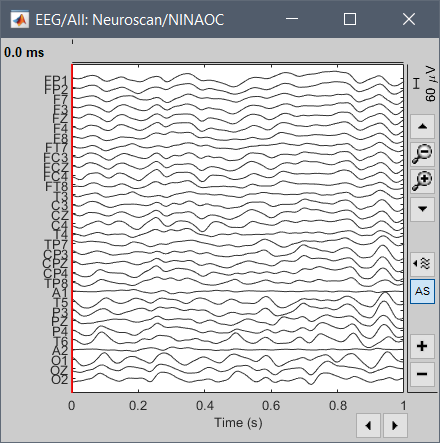

Thanks for your reply and sorry about the delay,
well I did what you suggest but It is still not working, in EDF browser the Fit to Pane option actually shows the signals but it changes the scale, I tryied to import the exported edf file again to Brainstorm but it was flat again, maybe it is just a local problem (like my computer or something like that) or a Neuroscan format specific issue.
Thanks
The screen captures I posted previously were generated with Brainstorm from the file you sent.
You should be able to reproduce exactly these figures with any recent version of Brainstorm.
- Link your file (ninaoc.cnt) to the Brainstorm database with menu "Review raw file"
- Right-click on the "Link to raw file" > File > Export to file > EDF format
- Link the new file (eg. ninaoc_export.edf) to the Brainstorm database with menu "Review raw file"
- Right-click on the original CNT "Link to raw file" > Import in database > Time window: 0s-1s, disable all the other options
- Right-click on the exported EDF "Link to raw file" > Import in database > same options
- Open the two imported 1s-epochs side by side
Hi!
Thanks for the reply!
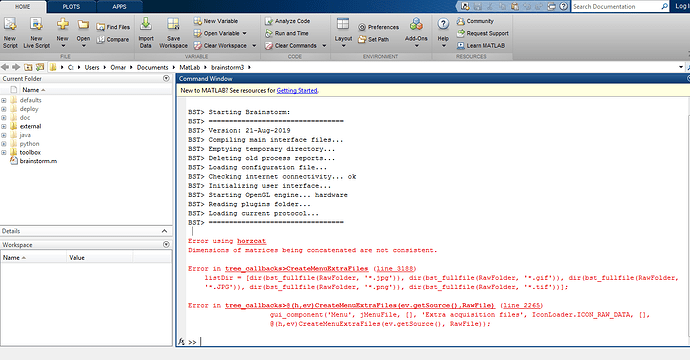
I tryied but everytime I Right-click over the file, I get this error message:
BST> Starting Brainstorm:
BST> =================================
BST> Version: 21-Aug-2019
BST> Compiling main interface files...
BST> Emptying temporary directory...
BST> Deleting old process reports...
BST> Loading configuration file...
BST> Checking internet connectivity... ok
BST> Initializing user interface...
BST> Starting OpenGL engine... hardware
BST> Reading plugins folder...
BST> Loading current protocol...
BST> =================================
Error using horzcat
Dimensions of matrices being concatenated are not consistent.
Error in tree_callbacks>CreateMenuExtraFiles (line 3188)
listDir = [dir(bst_fullfile(RawFolder, '.jpg')), dir(bst_fullfile(RawFolder, '.gif')), dir(bst_fullfile(RawFolder,
'.JPG')), dir(bst_fullfile(RawFolder, '.png')), dir(bst_fullfile(RawFolder, '*.tif'))];
Error in tree_callbacks>@(h,ev)CreateMenuExtraFiles(ev.getSource(),RawFile) (line 2265)
gui_component('Menu', jMenuFile, , 'Extra acquisition files', IconLoader.ICON_RAW_DATA, ,
@(h,ev)CreateMenuExtraFiles(ev.getSource(), RawFile));

Hello
I don't understand the origin of this bug...
I added some extra checks in this function: https://github.com/brainstorm-tools/brainstorm3/commit/f25d73124dd163364a8c3c84a0414aa3f000c5cb#diff-2ef654c5427b5793045f26aba06dfc8a
Please try the following:
- Update Brainstorm (from the menus in the main Brainstorm window)
- Reload your subject (right-click on the subject in the Brainstorm database explorer > Reload)
- Try again clicking on "Export to file"
- If it doesn't work, delete the subject and create a new one. Try again.
If it doesn't work, could you try doing some extra debugging?
- Put a breakpoint in function tree_callbacks.m at line 3191 (the line that crashes) : click on the dash next to the line number to set a breakpoint
- Try to reproduce the error (right-click on the Link to raw file > File > Export to file)
- When the debugger stops, you can explore the contents of the variables. Please execute the following lines in your Matlab command window and copy-paste here the output you obtain for each of them:
>> RawFolder
>> bst_fullfile(RawFolder, '*.jpg')
>> dir(bst_fullfile(RawFolder, '*.jpg'))
>> [dir(bst_fullfile(RawFolder, '*.jpg')), dir(bst_fullfile(RawFolder, '*.gif'))]
>> listDir = [dir(bst_fullfile(RawFolder, '*.jpg')), dir(bst_fullfile(RawFolder, '*.gif')), dir(bst_fullfile(RawFolder, '*.png')), dir(bst_fullfile(RawFolder, '*.tif'))];
Thanks
Francois
Hi!
I made some test and realized that this bug is just when I use the file I uploaded (ninaoc.cnt), with another neuroscan files there is no error message, maybe the file I made is corrupt, something it happens, let me do more tests to compare results,
Thanks!