Hello everyone,
I have a couple questions about exporting EEG files to EDFs:
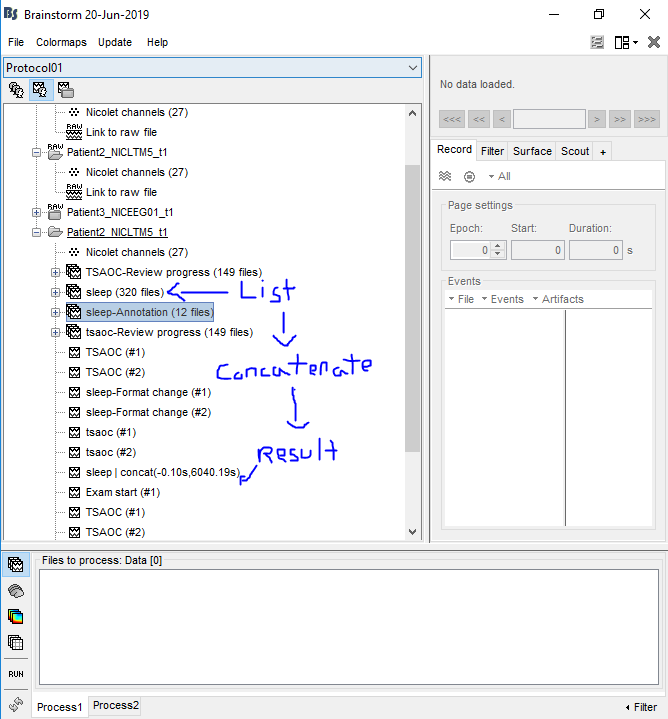
- The .e files essentially have segmentations like this:
[-bckg-][seg1][bckg][seg2][bckg]
When I use the concatenate time options on the files (to combine them into one so I can export it to an EDF), is it preserving the background signal,
or is it simply giving me:
[seg1][seg2] ?
- Am I exporting the files properly? The steps I take to export an .e file is:
-
Create a subject (this is where I place all my EEG files)
-
Right-Click > Import MEG/EEG > Select my .e File > Select all the data from the sleep list and process them using concatenate time > Right-Click the newly generated Data > File > Export to File > EDF
Is there a better way to do this?
P.S I uploaded some pictures to give you a better understanding of what I meant by "sleep list" and "newly generated data"
Thank you so much for time!
-Shmyrde Jean-Paul