Hi all.
I am working on 5-min resting state recordings. I want to export time-frequency (Hilbert transform) as .nii including time (4D).
Here is my pipeline:
- Resampling (500 Hz)
- Band-pass filtering (0.1 - 90 Hz)
- T1 coregistration
- Artifact cleaning (visual inspection, ICA)
- Noise covariance from empty room recording
- sLoreta source modeling (overlapping spheres for head model modeling)
- Hilbert transform (using scouts, grouping in bands, measure as power, 1/f compensation).
- Export volumes (4D, volume downsample=2).
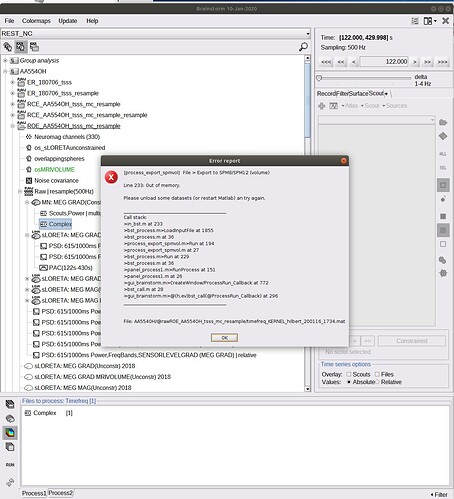
At this point I receive the error "Index exceeds array bounds". I already tried to further downsample the raw file or the TF maps, to check matlab preferences (I unchecked the box for array limitations), to reduce the number of vertices of the cortex, to change scout atlas and different preferences in the export window, without success.
Do you have any suggestion? I am not able to recognize the problem.
Thanks a lot for the support in advance.
Angelantonio
I am working on 5-min resting state recordings. I want to export time-frequency (Hilbert transform) as .nii including time (4D).
Are you planning on studying the evolution of the power by frequency band during these 5min?
Otherwise you might not need the time dimension in your analysis, and might be interested only in power by frequency band, as illustrated in this tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/RestingOmega
Band-pass filtering (0.1 - 90 Hz)
If your goal is to do some frequency or time-frequency analysis, you probably don't need to apply any frequency filter before.
At this point I receive the error "Index exceeds array bounds".
I'm not sure why you get this error. I've tested this with the tutorial dataset and it works as expected.
Please post the full error message, together with screen captures of the database explorer showing the file you are trying to export and the options for the export process.
Thanks Francois for the answer.
1)Yes, I need the evolution of the power by frequency during time ( I want to export the data to compute an ICA on them).
2)I did it trying to reduce the weight of the data, but I know it's not necessary since hilbert transform already apply filters;
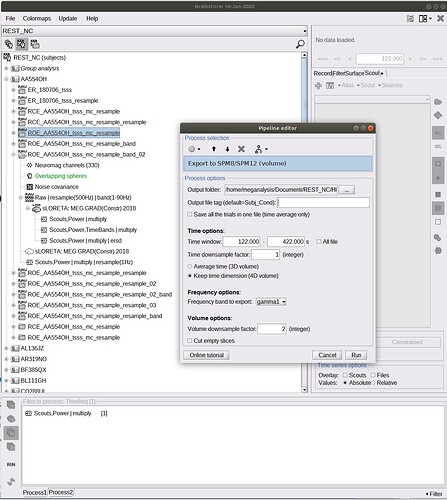
3)These are the screens:
Do you think it may be due to the specifications of my workstation?
Thanks for the support.
Angelantonio
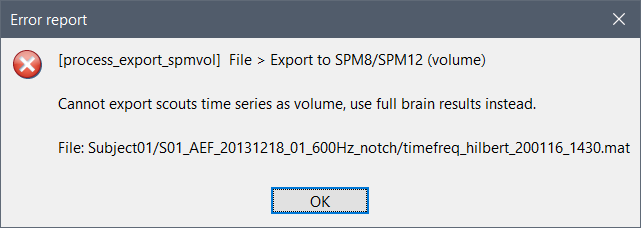
The name of the file in your database shows you have computed the time-frequency decomposition with Hilbert transform only on a few scouts. There is no spatial information in your file anymore, just the TF maps for a few scouts signals. You can't export this as a 3D or 4D volume.
Either you work only with these signals, or you recompute the TF for the full volume (which would create gigantic files you probably can't process with your computer).
I added an explicit error message to avoid future confusion:
Bug fix: Add error message when trying to export scouts TF · brainstorm-tools/brainstorm3@0318281 · GitHub
Hello Francois!
Thanks for the help. I compute TF on scouts because I had errors computing on full volume.
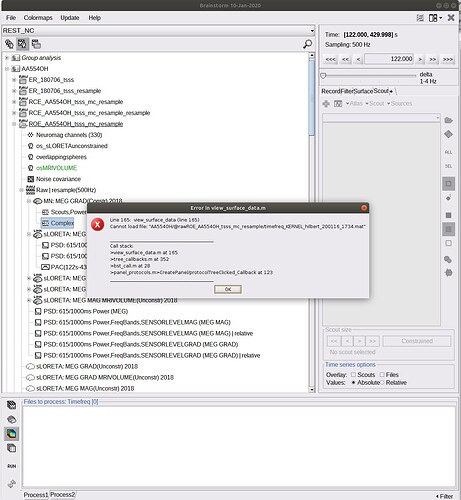
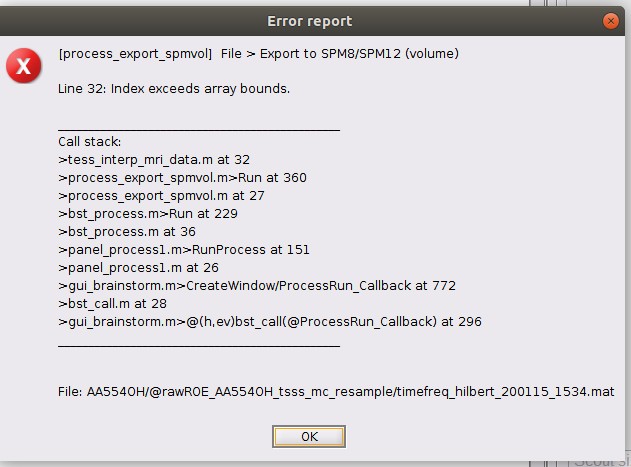
Yesterday I tried to compute TF for the full volume after reducing the cortex to 3002 vertices. Brainstorm created a big file (2.7 gb) that I am not able to open: these are the error windows.
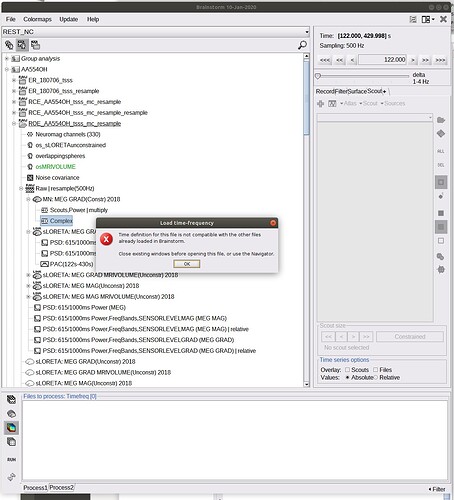
I tried also to export the TF (full volume), with no success:
If you think the problem depends on the computer specifications, have you got some suggestion about:
- specifications needed for this kind of analysis
- a way to reduce the amount of data, without losing too much information?
Kind regards,
Angelantonio
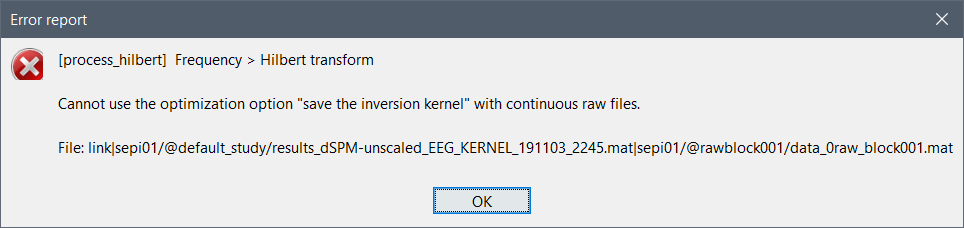
The option "Save inversion kernel" is not compatible with continuous files, but this was not caught previously. I added an error message to prevent this from happening again:
Bug fix: Add error message when using the option "save kernel" with c… · brainstorm-tools/brainstorm3@e900e92 · GitHub
If you want to use this option, you need to import the recordings first to the database.
If you didn't use this option, then the file generated is probably too big to be loaded in memory and displayed.
The time-frequency processes are not meant to be executed on long continuous recordings, but rather on short segments on which you want to follow the fine temporal dynamics.
I did it!
I downsampled the recording to 250 Hz and imported a time interval to the database, I also downsampled the cortex to 1000 vertices. Then I was able to compute the Hilbert on full volume. In this way I was able to manage the data.
Thanks a lot for the support.
Angelantonio