Hello,
I have an EEG recording in .rhs format (Intan), which is very large (11Go, 20kHz) and contains the values of 4 different subjects (mice). I was able to import the signal in a subject folder, but not extract specific channels into 4 different subject folders. Is it possible to do it in the Brainstorm GUI ?
Thanks for your advices
You can extract the individual channels from the imported file by running the process Extract > Extract values for each channel, this will create a matrix (![]() ) file with the data for the channel.
) file with the data for the channel.
![]() Note, that at this point each matrix file (time series of a given channel) is located in the same folder, as they are derivatives of the same recordings file (
Note, that at this point each matrix file (time series of a given channel) is located in the same folder, as they are derivatives of the same recordings file (![]() ) associated with a channel file (
) associated with a channel file (![]() ).
).
If you want to have a separated recordings for each channel, after extracting the time series for each channel, right-click on the resulting matrix and select Review as raw this will create a raw recording (![]() ) with data of only that channel
) with data of only that channel
Thank you Raymundo for your answer, but I do not have access to the "Extract values" option (greyed out), if I try to apply it to my data, I have the following error:
"Error : Data type mismatch.
Invalid inputs for process:
"Extract values" "
Have you ever tried to apply this extract option to .rhs file ?
It may be grayed out as you are working on the raw recording (![]() )
)
Have you imported the raw recording first? You need to import the data first.
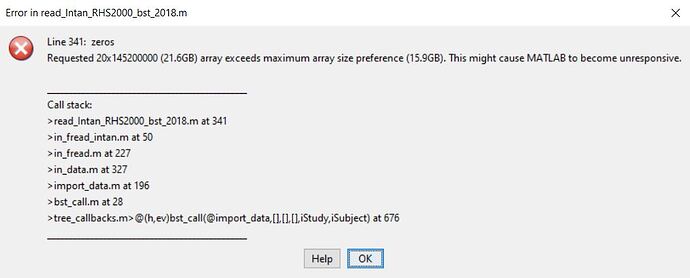
Thanks for your answer but I can't import my recording because of its size, it produces a Matlab error in 'read_Intan_RHS2000_bst_2018.m'
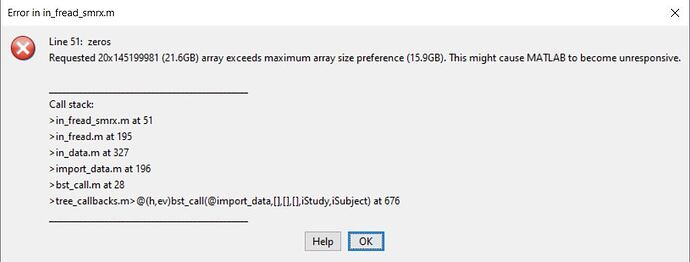
I tried another way: I imported my .RHS recording into Spike2, converting it in a .SMRX file, and I downsampled it from 20kHz to 1kHz. And then I tried to import this downsampled SMRX file into Brainstorm but I still have the same Matlab error but in the in_fread_smrx.m function, it seems files that can be imported are limited in length.
The error message suggest that downsampling was not performed.
Indeed, my file keeps track of the raw signals despite the downsampling. I will try to use another intermediate format.
Thank you for your answers
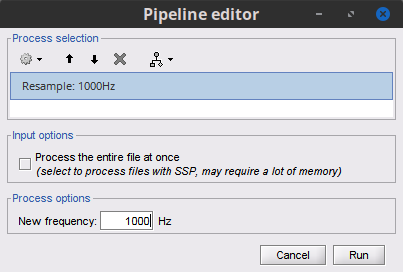
Note sure why you need an intermediate format, resampling can be performed directly on Brainstorm before importing the raw file. Once the raw data is linked to the Brainstorm database, you could resample it in Brainstorm, directly. Process Pre-process > Resample
Just do not check the checkbox Process the entire file at once, so you do not run into RAM problems.