Hi all,
I would like to retrieve the fiducials (nasion, lpa and rpa) coordinates from my MRI file in the anatomy folder using a script. However, I specifically need the coordinates in MNI.
These appear when I open the anatomt file in the gui viewer (highlighted in picture attached), but not when I load the .mat file from the anatomy folder. The only ones that I can find in that .mat file are the coordinates in MRI format. Does anyone know an easy/straighforward way to access these values with a script? (i.e. without having to read them from the gui?)
Thanks in advance,
Fran
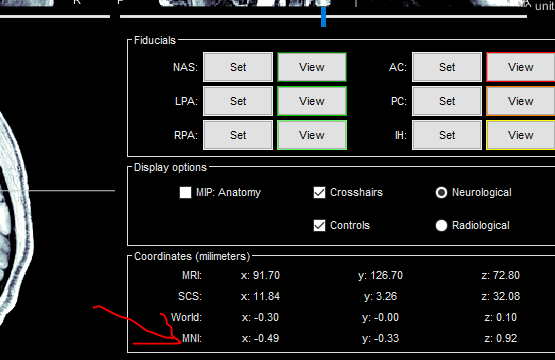
I would like to retrieve the fiducials (nasion, lpa and rpa) coordinates from my MRI file in the anatomy folder using a script. However, I specifically need the coordinates in MNI.
Does anyone know an easy/straighforward way to access these values with a script?
MriMat = in_mri_bst('C:\Work\Protocols\TutorialEpilepsy\anat\sepi01\subjectimage_MRI.mat');
NAS_mni = cs_convert(sMri, 'mri', 'mni', sMri.SCS.NAS ./ 1000) .* 1000;
LPA_mni = cs_convert(sMri, 'mri', 'mni', sMri.SCS.LPA ./ 1000) .* 1000;
RPA_mni = cs_convert(sMri, 'mri', 'mni', sMri.SCS.RPA ./ 1000) .* 1000;
The fiducials are saved in the MRI structure in millimeters while everything else in Brainstorm works in meters. Hence the 1000 factors.
https://neuroimage.usc.edu/brainstorm/CoordinateSystems#Converting_between_coordinate_systems
Thanks so much Francois, it worked. I only replaced "MriMat" by "sMri" in the first line.
