hello Francois,
I am having an issue that i think is related. I tried to do a script that is loding .eeg file (brainamp), filtering and resampling them and then export as .edf.
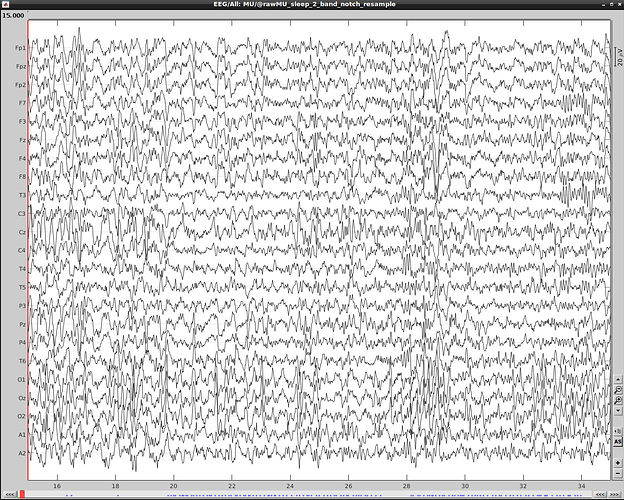
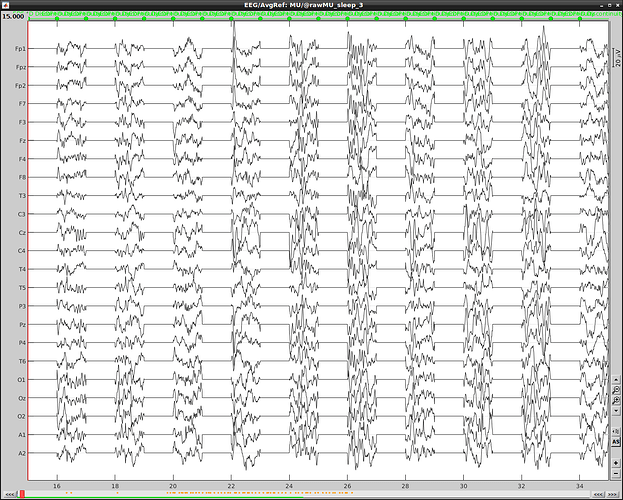
However, the created edf file doesn't correspond to the data in brainstorm and contains data discontinuty:
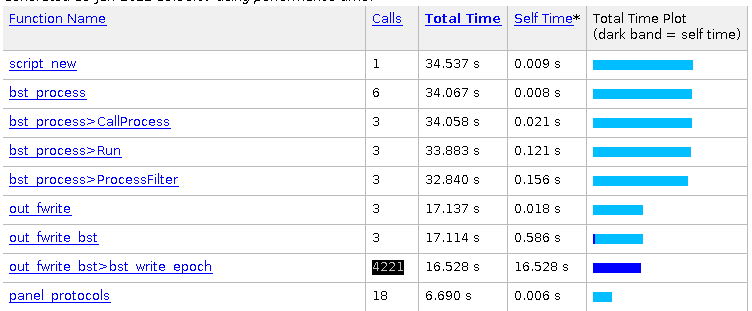
File in brainstorm (after resampling):
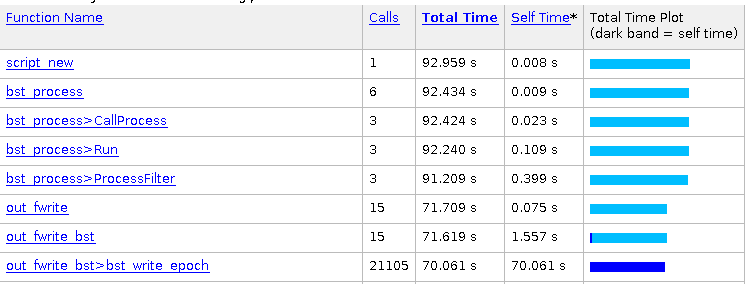
After exportation:
Brainstorm is telling me: WARNING: Found discontinuity between 1389.000s and 1390.000s, expect blank data in between.
WARNING: Found discontinuity between 1390.000s and 1391.000s, expect blank data in between.
...
The script i used:
% Script generated by Brainstorm (28-Jan-2022)
data_folder='/NAS/home/edelaire/Documents/data/normal_sleep/';
export_folder = '/NAS/home/edelaire/Desktop/marking';
%%
if ~brainstorm('status')
brainstorm
end
bst_report('Start');
protocol_name = 'For_laure';
if isempty(bst_get('Protocol', protocol_name))
gui_brainstorm('CreateProtocol', protocol_name, 1, 0); % UseDefaultAnat=1, UseDefaultChannel=0
end
subjects = dir( fullfile(data_folder,'*')); % ici on veut lister uniquement les repertoires sujets et pas leur contenu
for i_subject = 7:length(subjects)
% Creation du sujet
sSubject = bst_get('Subject', subjects(i_subject).name, 1);
if isempty(sSubject)
[sSubject, iSubject] = db_add_subject(subjects(i_subject).name, [], 1, 0);
end
EEG_recordings = dir( fullfile(data_folder, subjects(i_subject).name,'EEG/*.eeg')); % ici on veut lister uniquement les repertoires sujets et pas leur contenu
for i_recordings = 1:length(EEG_recordings)
sFiles = bst_process('CallProcess', 'process_import_data_raw', {}, [], ...
'subjectname', subjects(i_subject).name, ...
'datafile', {fullfile(EEG_recordings(i_recordings).folder,EEG_recordings(i_recordings).name), 'EEG-BRAINAMP'}, ...
'channelreplace', 1, ...
'channelalign', 1, ...
'evtmode', 'value');
sFiles = bst_process('CallProcess', 'process_bandpass', sFiles, [], ...
'sensortypes', '', ...
'highpass', 0.5, ...
'lowpass', 100, ...
'tranband', 0, ...
'attenuation', 'strict', ... % 60dB
'ver', '2019', ... % 2019
'mirror', 0, ...
'overwrite', 0);
% Process: Notch filter: 60Hz
sFiles = bst_process('CallProcess', 'process_notch', sFiles, [], ...
'sensortypes', '', ...
'freqlist', 60, ...
'cutoffW', 5, ...
'useold', 0, ...
'overwrite', 0);
% Process: Resample: 250Hz
sFiles = bst_process('CallProcess', 'process_resample', sFiles, [], ...
'freq', 250, ...
'overwrite', 0);
[filepath,name,ext] = fileparts(EEG_recordings(i_recordings).name);
if ~exist(fullfile(export_folder,subjects(i_subject).name))
mkdir(fullfile(export_folder,subjects(i_subject).name))
end
[ExportFile, sFileOut] = export_data( sFiles.FileName, [], fullfile(export_folder,subjects(i_subject).name,[name '.edf']) );
end
end
Edit:
it seems that changing the way i import the data from continous file to recording in the db if fixing the issue:
Changing :
sFiles = bst_process('CallProcess', 'process_import_data_raw', {}, [], ...
'subjectname', subjects(i_subject).name, ...
'datafile', {fullfile(EEG_recordings(i_recordings).folder,EEG_recordings(i_recordings).name), 'EEG-BRAINAMP'}, ...
'channelreplace', 1, ...
'channelalign', 1, ...
'evtmode', 'value');
to
sFiles = import_data(fullfile(EEG_recordings(i_recordings).folder,EEG_recordings(i_recordings).name),[], 'EEG-BRAINAMP', [], iSubject);