Dear All,
I'm going through the tutorial 3: once I input the fiducial coordinates and save I don't see any import from FreeSurfer and I don't see the same anatomy as the one displayed in the Tutorial.
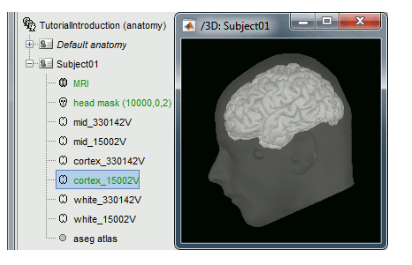
I only continue seeing the same as after the import and I was under the impression that such data was already imported from FreeSurfer, is it not? Or do I need to install FreeSufer first?
And indeed, I have tried to install FreeSurfer and not sure if you can help with FreeSurfer installation but I have had absolutely no success with it!!! And any help would be welcome!
I have installed FreeSurfer in Virtual Box (4096 MB Memory; 32 MD Video Memory; Ubuntu 64 Bits) with an Ubuntu system that works fine but I cannot unzip the “freesurfer-Linux-centos6_x86_64-stable-pub-v6.0.0.tar.gz” file to install FreeSurfer and being new to Linux:
i. I’m not sure if at all the files are unzipped from the .gz file.
ii. And I have been unable to create the Bash files to define the environment for it.
Any help appreciated!
Kind Regards,
Rodrigo Montenegro
