Hello brainstorm, I am a newbie so I apologize if this is a trivial question.
I also apologize if the topic is not the right one.
I upload a code that should do a source reconstruction analysis for a few subjects iteratively.
I have their eeg recordings ( .edf format ) located in "ifilepath", but I do not have any kind of anatomy so I wish to use the default one (line 21-31). Moreover I would like to use my own channel location file called "STD64.sfp" (line 53-57).
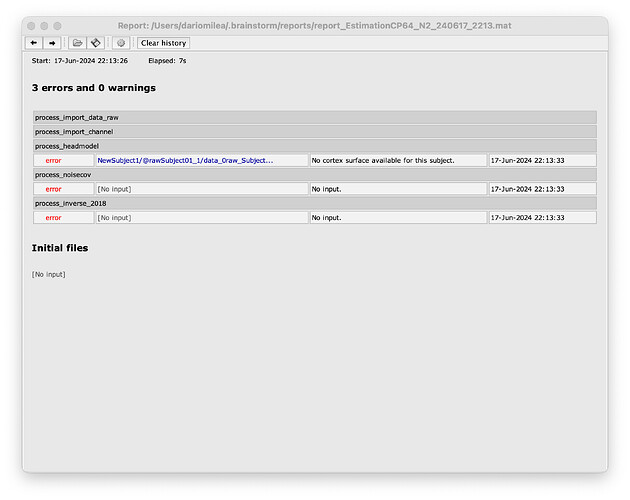
My questions are: (1) when I run the code a window opens for choosing the channel location file; maybe line 53-57 are not well written? (2) Then, an error occurs in the process_headmodel since there is no cortex surface for the subject (see the figure)
test.m (3.6 KB)
However, if I create a new protocol by GUI using the default anatomy (of course I erase lines from 10 to 31 of the code) the error does not occur (in this case a window open for choosing the reference MRI).
At the moment I am using this alternative but how can I fix the script without the use of the GUI?
Thank you very much.
If you are complete new to to Brainstorm, we strongly suggest you to reading all the introductions tutorials (section "Get started"), using the example dataset, in order to learn how to use the software. This will also give you the jargon you would need.
https://neuroimage.usc.edu/brainstorm/Tutorials
If you haven't take a look to the scripting tutorial:
Moreover, Brainstorm distribution includes scripts that reproduce the different tutorials (in folder brainstorm3/toolbox/scripts).
Try first to perform the actions from your pipeline in the GUI, then proceed to script it.
About your script:
-
Never call clear if Brainstorm is running, it will clear variables, functions and classes that are needed in Brainstorm
-
The call to process_import_anatomy is incorrect, it expects a folder or file with pre-processed individual MRI. As you want to use the Default anatomy, create a Subject like this:
[sSubject, iSubject] = db_add_subject(SubjectName, [], 1, 0);
-
The error in process_headmodel is the result of an invalid anatomy. Because it was not created in the first step.
Thank you very much for your fast reply.
It seems to me there are no problems from GUI.
Thank you