Dear Sylvain, dear Francois,
is there a way to import surfaces meshes made by fsl into brainstorm?
Regards,
Marcel
Dear Sylvain, dear Francois,
is there a way to import surfaces meshes made by fsl into brainstorm?
Regards,
Marcel
Dear Marcel,
Aren’t FSL and FreeSurfer using the same file formats for the surfaces?
What happens if you try to open your surfaces selecting the “freesurfer” file format in the dialog box?
If there are surface formats that are not supported, provide me with a Matlab function (GPL) that reads them, and I will integrate that in the Brainstorm distribution.
Cheers,
Francois
Dear Francois,
FSL saves the meshes in nifti. Reading nifti is already implented in brainstorm for MRI scans. Maybe it is also possible to use this funtion to read meshes. Thank you for your help!
regards,
Marcel
Dear Marcel,
NIfTI is a file format used to store 3D/4D volumes, not meshes.
If you provide me with the appropriate information (short description of the format, matlab function to read it, example files), I will add the support for the format you need.
Cheers,
Francois
Dear Francois,
Of course you are right! Interestingly fsl saves the meshes in two different formats: the brain mesh is saved in vtk-format. The m-file, which can read vtk should be here. http://www.mathworks.com/matlabcentral/fileexchange/5355-toolbox-graph/content/toolbox_graph/read_vtk.m. The skull meshes are in files, which end with .off, but I don’t know this format and unfortunately I can’t find a reader.
Best regards,
Marcel
The off format is made by Geomview http://www.geomview.org/ , but I could not find a matlab reader.
Best regards,
Marcel
Dear Francois,
the reason to use fsl is, that freesurfers usually needs about one day to process one brain and the automatic pipepline does not include segmentation of the hippocampus, which is very important for temporal lobe epilepsy. BrainVISA runs very unstable. I had many problems in brain segmentation. As far as I know fsl segmentation runs fast, it is user friendly and it produces acceptable results. If there are no other persons interested in using fsl for segmentation, maybe it is too much work to implement it into brainstorm.
Thank you for your interest and your efforts!
Marcel
Hi Marcel,
I added the support for meshes created with FSL:
But the results are terribly ugly:
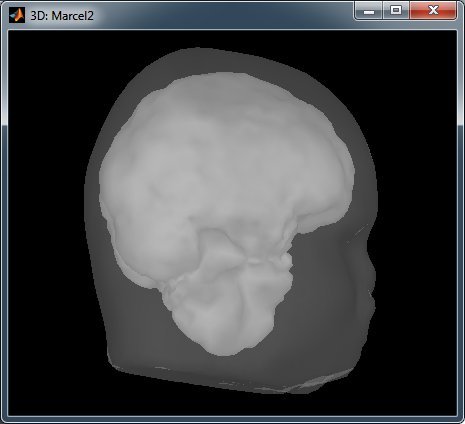
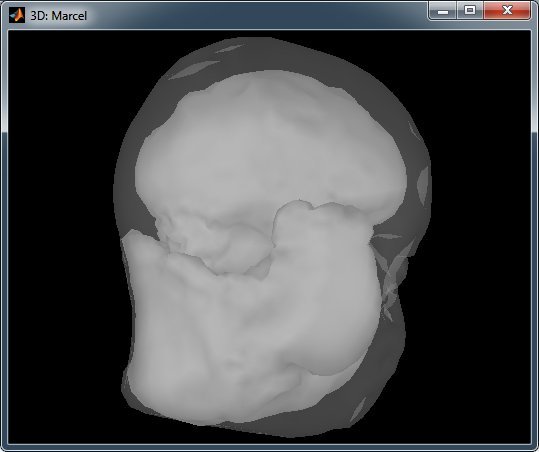
Cheers,
Francois
Hi Francois,
sorry for the atrocious segmentation results using fsl. I could not spend to much time to test it by myself, which I should have done before sending it to someone else. A colleague of mine, who is working on finite element head modelling, is using fsl as one of his segmentation tools. I just wanted to find an alternative for freesurfer or brainvisa. There might be the chance that I’m not using it right.
Marcel
Hi Francois,
Is there any way to “[I]Import anatomy folder[/I]” generated by [B]FSL[/B] into the Brainstorm? I tried to do it using the “FreeSurfer” option but didn’t work out.
We are trying to work with infant MRI data pre-processed by FSL for infant EEG source estimation.
Please help.
The data description is available here: http://jerlab.psych.sc.edu/NeurodevelopmentalMRIDatabase/description.html.
And, I am copying it below:
Description.
The database consists of MRI average templates for a number of ages; in 1-3 month increments through 18 months; then half-year increments through 19-5 years; then 5 year increments through 89 years. The templates were done separately for brain and head. Also included are segmentation PVE volumes for gm/wm/csf; T2W-derived CSF; and non-myelinated axons (NMA) for infants. Access to the dataset is separated by ages (infants; 0-12 mo; preschool, 15 mo through 4-0 years; children 4-5 through 10-5 yrs; adolescents 11-0 through 17-5 yrs; adults 20-89 years).
The segment data for ages 15-months and older consists of GM, WM, CSF, and T2W-derived CSF. The best combination of segments would be the image_aposteriori_seg data, using GM, WM, and T2W-derived CSF for priors. For 3 through 12 months, the best combination of segments would be the nma_seg data; using GM, WM, NMA, and T2W-derived CSF. The “CSF” PVE segments are “Other Matter” in a 3-class segmentation (GM, WM, “Other Matter”) and does not reflect actual CSF. The T2W-derived CSF is identified as bright voxels in the T2W scan and represent actual CSF in the brain or head. There is an atlas derived from FSL “Harvard-Oxford” cortical and subcortical atlas for the infants, 8 10 12 14 16 18, and 20-24 year old templates.
Overview:
ANTS…brain.nii.gz: Average MRI template derived from extracted brain
ANTS…head.nii.gz: Average MRI template derived from whole head
ANTS…brain-head: brain extracted from head template
ANTS…T2W_brain: MRI template separate for extracted brain T2W
ANTS…T2W_head: MRI template separate for whole head T2W
Segments
AVG…T2W_brain…: T2W for individual participants, warped to template, averaged
AVG…image_seg_…: Image-based segment averages
AVG…image_aposteriori_seg_… : Age-template priors with a posteriori FAST
AVG…MNI_aposteriori_seg_…: AVG of MNI-template priors, with a posteriori FAST
AVG…nma_seg_: For infants, non-myelinated axons separate from gray matter
AVG…seg_csf: “Other matter” in 3-class segmentation
AVG…seg_t2wcsf: T2W-derived CSF
Atlas:
ANTS…brain…brainstem: The individual files have the brain areas
ANTS…brain_atlas: Segmented atlas for all brain areas
Hello,
There is currently no support for the results of the FSL segmentation.
I tried to add this a while ago and remember that there was too much missing information for me to keep on working with it. But I probably didn’t spend enough time to look for all the possible information…
I don’t have time for implementing this right now, but I could help someone to do it. The idea would be to produce a script import_anatomy_fsl.m, equivalent to the other import_anatomy_* scripts for FreeSurfer, BrainSuite and BrainVISA. The output from those three programs is well documented on the tutorial page. Maybe the FSL developers would be interested in contributing to this.
Cheers,
Francois
Thank you Francois.
I will explore the scripts for importing.
Regards,
Berdakh.
Hello Francois,
I would really like to work on implementing the Brainstorm interface for FSL. Could you please guide me on this?
Thanks,
Berdakh.