Hello Brainstorm Community,
I have 10 participants each with 5 min resting-state EEG data in an eyes-closed condition.
Each participant went through 3-shell sphere forward modeling via cortex surface source space with no-noise covariance computation and constrained sLORETA minimum norm imaging.
I need a grand-average source-level brain image across the 10 participants in their frequency bands along with the corresponding power spectrum density (PSD) data for the bilateral superior temporal gyri.
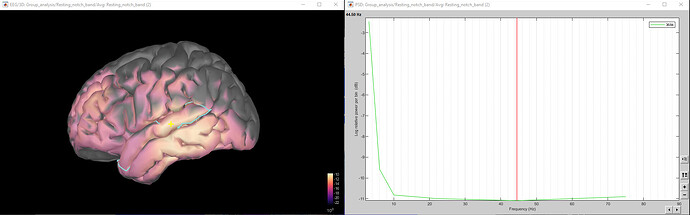
The below-left image is the grand-average source-level brain image across 2 participants at the gamma1 (30 - 59 Hz) band and the below-right image is the resulting PSD of the yellow cross at the left superior temporal gyrus.
What I really need is to be able to select the entire left and right superior temporal gyri (not selecting through the yellow cross but using scouts that select the specific brain region) and include both PSDs into a single graph that can also show the changes of the topography at the source level across the different frequency bands from delta to gamma2 bands.
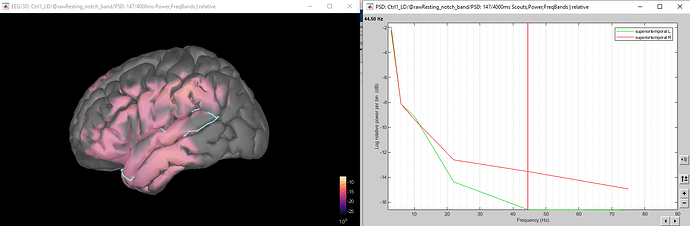
The image below shows what this could potentially look like for a single participant - I just need to be able to do this at the grand-average level (which I haven't found out the method for yet).
How can I achieve this?