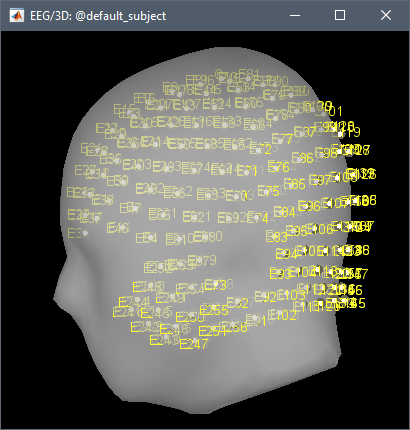
I do not know the exact difference between GSN hydricel 256 E001 and GSN hydricel 256 E1?
Also, I do not know what is the relationships between these two template with the following link. It looks you used some normalization, could you explain what is the normalization?
I do not know the exact difference between GSN hydricel 256 E001 and GSN hydricel 256 E1?
The naming of the electrodes, e.g. "E001" vs. "E1".
Different versions of the NetStation (or different versions of the caps) used different conventions.
It looks you used some normalization, could you explain what is the normalization?
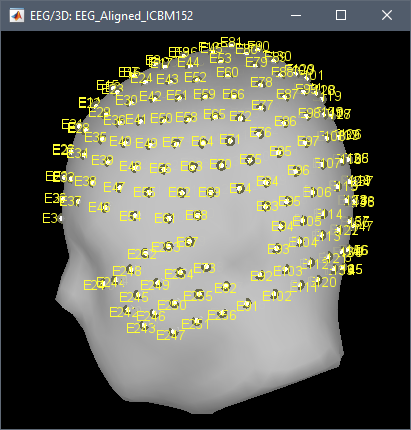
The default 3D positions provided by EGI do not correspond to any standard anatomy template.
We manually adjusted the positions of the electrodes provided by EGI on the head surface of the MNI ICBM152 template.
I wanna normalized the MRI system (MNI) to an EEG system (GSN hydricel 256 E001 or GSN hydricel 256 E1), and I know how to do it, but I do not know what is the the transformation matrix that you used. Do you think it is possible for you to explain how to transfer these two systems? which articles do you use?
Do you think it is possible for you to explain how to transfer these two systems? which articles do you use?
This is a manual transformation, validation is done only visually.
EGI used the head of one particular individual for digitizing the default positions of the various caps. This has no value as a standard, do not expect to find a more formal solution.
Additionally, the HydroCel nets are very extensible: put them twice on the same head, you get different positions for most of the electrodes. You can't expect to get reproducible positions for a given EEG net. You can't expect to get a precision better than 1cm or 2cm when using template 3D positions.
I wanna normalized the MRI system (MNI) to an EEG system (GSN hydricel 256 E001 or GSN hydricel 256 E1)
To convert between coordinates systems (e.g. Brainstorm SCS and MNI), see the tutorials:
Thank you Francois! that is great.
I have two quick question. I have MRI images (NIFTI file), and I want to change every voxel of this nifti file to EEG coordinate. Is there any function to do it in Brainstorm? because I know that I could use the transfer function of cs_convert (voxel to SCS) and after that I apply it to the mesh of the nifti file, but I wanna know that is there any function to do this? (Generally my work is that align my nifti file to the EEG Cap, and extract both of them for my future work.)
Also, another question is could I extract the mesh of the EEG cap from brainstorm too? I am just thinking that for showing the EEG Cap, the software apply a marching cube algorithm. I wanna know that this file also available for use or not?
Thank you for your time,
I'm sorry, I'm not sure I understand what you need beyond cs_convert.
I'm not sure either what you call the "EEG coordinate". Is it the Brainstorm SCS?
could I extract the mesh of the EEG cap from brainstorm too? I am just thinking that for showing the EEG Cap, the software apply a marching cube algorithm. I wanna know that this file also available for use or not?
What EEG cap mesh?
If you want to access the patch object created when you double-click on the channel file, you can get directly from the Matlab figure:
hPatchEeg = findobj(gcf, 'Type', 'patch', 'Tag', 'SensorsPatch');
Faces = get(hPatchEeg, 'Faces');
Vertices = get(hPatchEeg, 'Vertices');
Thank you for your response Franc,
Yes, I mean the Brainstorm SCS, I wanna transfer every voxel of my nifti file (MRI images) to Brainstorm SCS. Is there any function to do that?
And also thank you for your second answer. I got it. Thanks!
You can use the approach that is used in the MRI reslicing function, which defines the list of the coordinates XYZ for each voxel:
And then call cs_convert to obtain SCS coordinates: