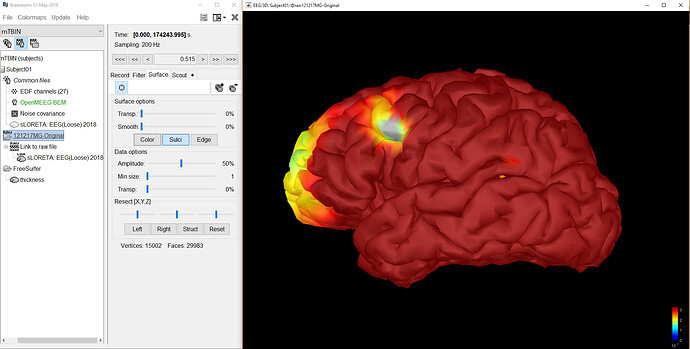
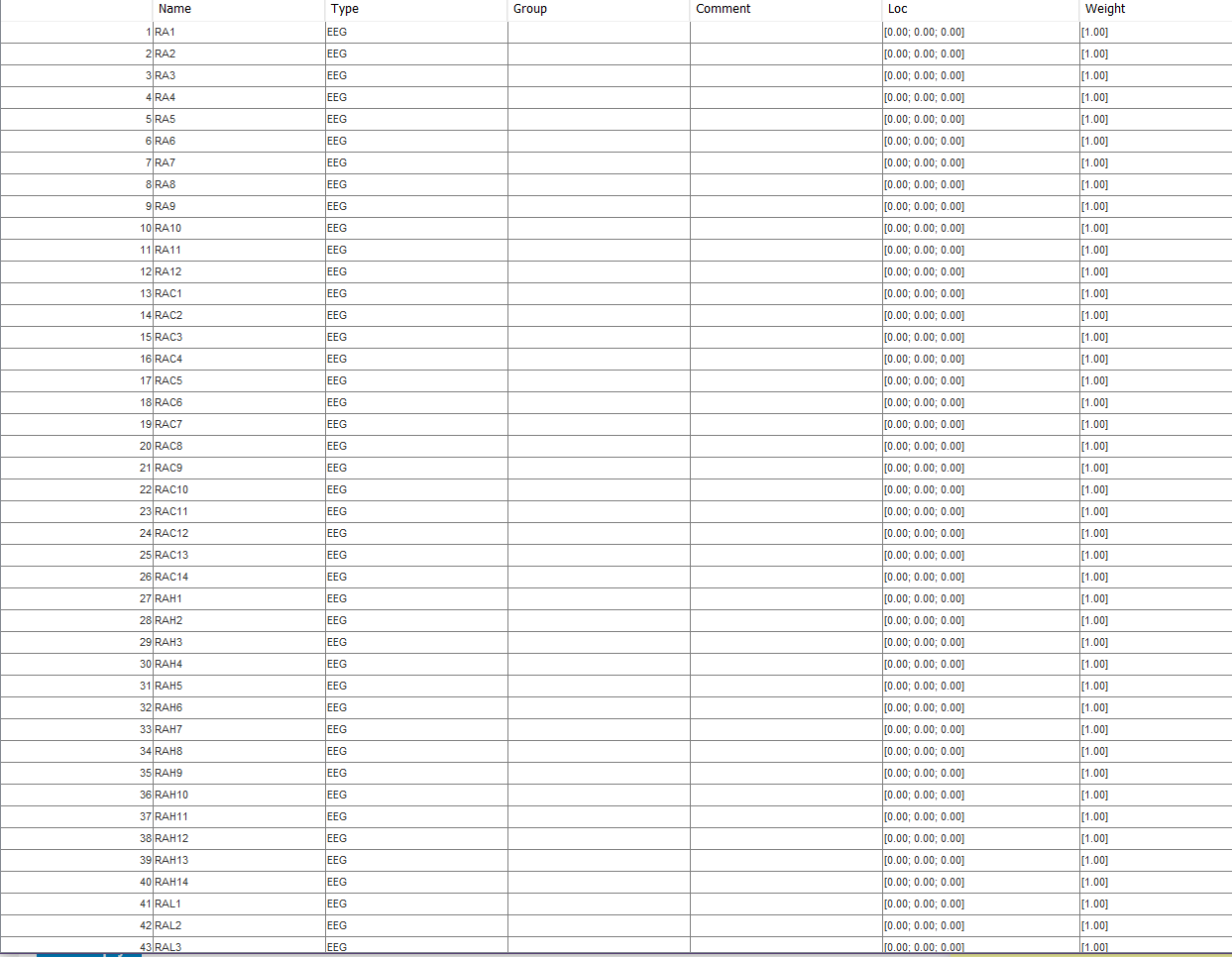
I am working with a single subject and 48-hour EEG recordings. I have successfully imported the anatomy from the mri and freesurfer pipeline. I exported the EEG data from a proprietary .ieeg format to .edf and it seems reasonable.
Although my subject is a head injury, I am following the EEG and Epilepsy tutorial, since I didn't see any others specifically for EEG.
I am stuck on the "Source Analysis: Surface" step in preparation for projecting the EEG data onto the surface model. I successfully generated the BEM surfaces, but when I click on the "Compute head model" from the functional menu, I get a popup "No valid sensor types to estimate a head model".
Here is my channel list (not sure if the zeros in the loc column are normal or an artifact from a bad export to edf):
Are there any suggestions to help get things unstuck?
Kind regards,
-Matt
Hi Matt,
You need to define 3D positions for your EEG electrodes, in order to reconstruct the source activity on the brain surface. If you have realistic positions (eg. digitized with a Polhemus, like in the EEG/Epilepsy tutorial), or use standard 10-20 locations.
If you don’t have accurate 3D positions for the electrodes but still want to use the patient’s anatomy for the source estimation, you need to use standard positions and adjust them manually on the head of the patient.
- Right-click on the channel file > Add EEG positions > ICBM152 > Generic > 10-20
- Right-click on the the channel file > MRI registration > Edit: Read the tooltips for all the buttons in the toolbar to understand what they do, then adjust the position of the electrodes on the head (rotations, translations, automatic fit…), then project the electrodes on the head surface, and finally adjust manually the electrodes one by one if needed.
Cheers,
Francois
Hi Francois,
Thanks for the support. I added the EEG positions as you suggested but am unsure how to correctly perform the MRI registration. I had assumed that the "Add EEG Positions" would have placed the electrodes in a valid position, so for testing I just left them where they were as shown in this image:
However, when I went to Compute Head Model after Generating BEM surfaces, I got the following error:
I understand how to adjust the electrode positions on the head surface, but it seems like the ICBM152 Generic 10-20 placed them into the brain volume.
How do I know which electrodes are the troublemakers and how can I fix this?
Kind regards,
-Matt
Standard electrodes positions cannot fit the anatomy of you subject. Only you know how these electrodes were placed on the head, we cannot help you with this. You have to adjust manually the position of the electrodes the best you can, so that it looks more or less like the way the electrodes were placed on the subject’s head.
Read the help of the various buttons and follow this sequence:
- adjust the position of the electrodes on the head (rotations, translations, automatic fit…)
- project the electrodes on the head surface
- adjust manually the electrodes one by one if needed (if you think they are not where they are supposed to be)
Thanks very much for all of your help, the renderings look great. In looking at the cortical activations, is there a way to smoothly play the EEG recordings for several minutes at a time rather than clicking the sample/epoch advance button repeatedly? I would also like to be able to export recorded activations on the model as a video. Is that possible?
There is no « play » button, but it is not complicated to implement: write a for loop including a call to panel_time>SetCurrentTime, and adjust the speed with a sleep() call.
Or right-click on the 3D figure > Snapshot to create .avi movies.
Hello Francois,
I'm having a very similar problem that Matt was having and I, too, am getting the popup "no valid sensor types to estimate a head model"
These are some of my channels:
I tried right-clicking on the channel file and going to "add EEG positions" then going to ICBM152 then generic and then 10-20 and I got the popup "no channel matching the loaded cap"
I should probably add that I'm using FieldTrip channels and there are 108 (I'm pretty new to Brainstorm and just data analysis in general so I'm not 100% sure what I'm doing).
Any help would be greatly appreciated. Thank you!
Sincerely,
Arul
As explained in the tutorial "EEG and Epilepsy", that introduces this menu "Add EEG positions":
The channels are matched by name: the position file you import must include the labels of the electrodes and they must be named exactly in the the same way as in your recordings.
https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy#Prepare_the_channel_file
You channels are named RA1, RA2... None of the Brainstorm templates have such labels.
If you want to use default electrode positions for this electrode cap, you will need to provide your own position file. If your electrodes are following a 10-10 or 10-20 placement, you need to edit the channel names in your channel file, or create your own template with adjusted names.
I'm pretty new to Brainstorm and just data analysis in general so I'm not 100% sure what I'm doing
I would recommend you start by following the introduction tutorials at least until #19, then the advanced tutorial EEG and epilepsy, all using the example datasets provided.
https://neuroimage.usc.edu/brainstorm/Tutorials
Thank you so much! I will try that.