Dear brainstorm experts,
How can I add a volume parcellation to the CAT12 MRI Segmentation pipeline? I know there are several volume parcellation availables (AAL3, anatomy, cobra,...). However, I need to parcellate the T1w of one of my subjects using the first version of the AAL atlas (AAL1).
Could you please advise me on how to incorporate the AAL1 atlas into the segmentation pipeline? Are there any specific files that require editing?
Many thanks for your help
Cristóbal
Since the AAL1 and AAL2 atlas are already in the MNI space, it is possible to added them to any subject anatomy for which the MNI normalization was computed.
Right-click on the anatomy folder > Add MNI parcellation, you should see the MNI parcellations available for direct download Brainstorm. If you do not see AAL1, update your Brainstorm instance.
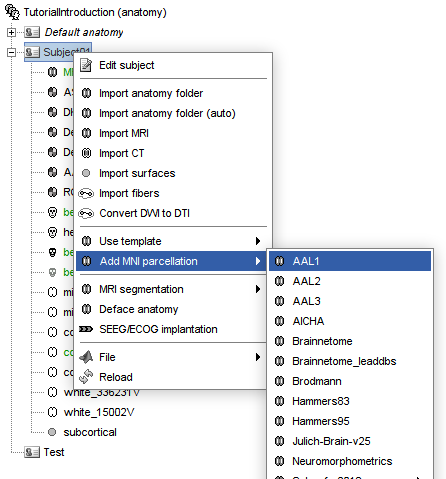
Dear Raymundo Cassani,
Thank you very much for your response. I have successfully applied the AAL1 atlas to my subject's anatomy. I would also like to ask clarification regarding how the software labels the voxels of the subject's T1w.
In the documentation, it states, "A non-linear deformation field can be applied to an MNI parcellation in order to create a volume parcellation in subject space" (https://neuroimage.usc.edu/brainstorm/Tutorials/DefaultAnatomy#MNI_parcellations). Does this process correspond to what I just did (project the atlas to my subject's anatomy)? Specifically, is the non-linear deformation applied to the atlas in order to align it with the subject's anatomy?
Could you please help me understand this statement better?
Thank you very much for your assistance.
Best Regards,
Cristóbal
Hi Cristóbal,
The documentation in the link is now updated for sake of clarity. " For a subject anatomy which MNI normalization was computed, the MNI parcellation in the subject space is obtained by reslicing the MNI parcellation into the subject space."
This is done with the Brainstorm function mri_reslice_mni()
brainstorm3/toolbox/anatomy/mri_reslice_mni.m at master · brainstorm-tools/brainstorm3 · GitHub
The "reslicing the MNI parcellation into the subject space" means that as the subject's anatomy is MNI normalized, each voxel in the subject anatomy is associated with a set (XYZ) of MNI coordinates, thus it is possible to get the label for that voxel according to the label for the same coordinates in the MNI parcellation. Thus, performing this for all the voxels in the subject anatomy, gives as result a MNI parcellation in the subject space.
This just indicates that if the MNI normalization was non-linear, computing the MNI parcellation in the subject space is also non-linear.
