Dear Brainstorm community,
I would like to construct EEG source space functional connectivity and perform group-level statistical analysis (BEM head model, wMNE method (constrained/unconstrained)). In the process, I have some questions.
-
According to my understanding, the sign of the time signal of each vertex in the source space is uncertain, i.e. flipping the sign of the source time signal and the corresponding row of the LeadField/ the column of ImagingKernel simultaneously doesn't change the result. Therefore, is it not applicable to construct functional connectivity with sign-related methods at the vertex level, such as Pearson correlation, etc... On the other hand, wPLI/PLI/PLV are not affected.
-
Due to the high time consumption and memory requirement of vertex-level functional connectivity construction, it is an intuitive way to reduce the dimensions of the time signals within the brain region. However, since the vertex signal can arbitrarily be 'flipped' by changing the sign, averaging the signal within the brain region (constrained model) or performing PCA to retain the first principal component seems to introduce unpredictable signal loss.
-
For the unconstrained model, making the analysis more complicated. If computing the connectivity matrix for each vertex for all the orientations (x,y,z), and taking the maximum connectivity measure across the three orientations. Then for connectivity between two brain regions, averaging the connectivity over all possible vertex pairs seems to be the most reasonable, but as with point 2, it is difficult for an ordinary computer to sustain such a large memory consumption. Reducing the dimensionality of the signal also faces the same challenge as in point 2.
-
The tutorial states that the "constrained" option is recommended for individual anatomy and the 'unconstrained' option is recommended for default anatomy. What is the reason for this? Can a constrained model achieve more accurate localization?
-
If only the alpha band is analyzed, is it equivalent to performing source localization and then filtering and filtering the original EEG data first and then performing source localization?
-
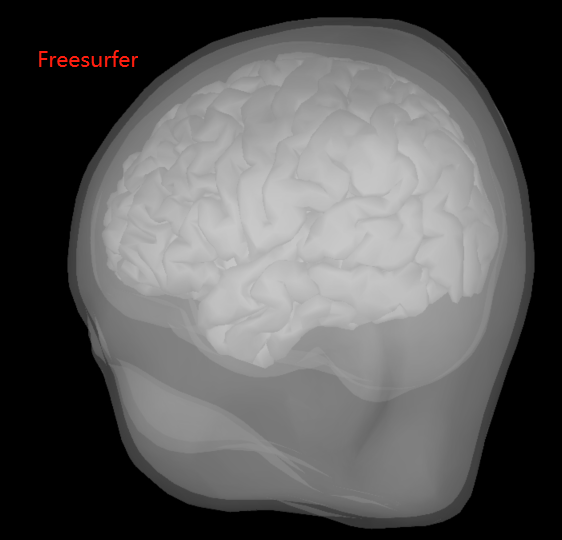
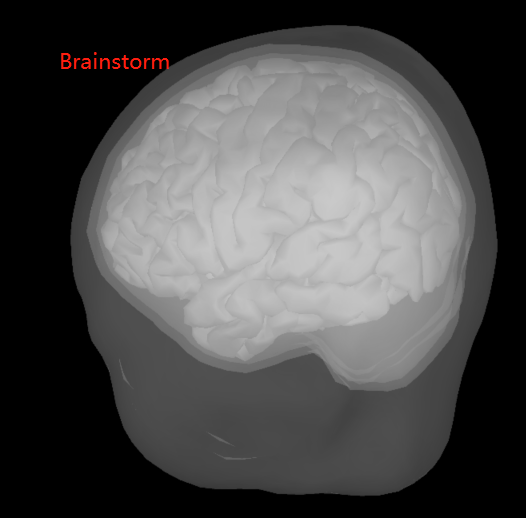
BEM surfaces based on Freesurfer and Brainstorm are significantly different. Which one is better to choose? (See the pictures below)
-
Does the Brainstorm team have any suggestions on the construction of the functional connectivity? For example, is it better to use constrained or unconstrained models, and if we want to calculate the functional connectivity at the brain region level, how to downscale the signals within the brain region?
Thank you so much!