Hi all,
I have processed my MRI via FreeSurfer:

But when I import the FreeSurfer Files into brainstorm, some erros happen:
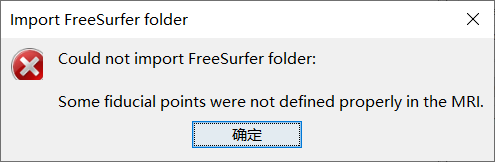
The tutorial as following:
But I still can not understand, What should I do?
Thanks so much!
Hi all,
I have processed my MRI via FreeSurfer:

But when I import the FreeSurfer Files into brainstorm, some erros happen:

The tutorial as following:
Thanks so much!
You should start by following the introduction tutorials, using the example dataset (all the tutorials in the section "Get started"):
https://neuroimage.usc.edu/brainstorm/Tutorials
Importing a FreeSurfer anatomy folder is explained in tutorial #2.
Thanks, Francois, one more question: Can brainstorm set fiducial points automatically (I have more than 500 subjects)? Or I need to check it one by one?
If you script your analysis, you can use the process "Import > Import anatomy > Compute MNI transformation" (equivalent of clicking on the button with the same name in the MRI Viewer).
This computes a 4x4 affine transformation to register the subject brain to the MNI template (ICBM152). If the anatomical fiducials (NAS/LPA/RPA) are not defined yet, this process also projects the default positions for these fiducials from the template to the subject anatomy.
Therefore yes, these points can be set automatically, at least approximately.
The problem is: how do you register the anatomy with the sensor positions?
Only the studies with no individual anatomical information (EEG with anatomy template + default electrode positions) can be ran fully automatically.
You can find various examples in the scripts corresponding to the tutorials in the section "Other analysis scenarios" (brainstorm3/toolbox/script/tutorial_*.m)
Perfect! Thanks so much Francois! Have a nice day