Hi Francois,
thanks for your response. I looked at the tutorial and it seems that it has what I need. However, when I tried to use the example to do what I need in my code, I get an error. Following is my code:
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% Script generated by Brainstorm (14-Apr-2015)
brainstorm nogui;
import_data;
% Input files
sFiles = {...
'NewSubject/Default/data_block001.mat'};
% Start a new report
bst_report('Start', sFiles);
% Process: Compute noise covariance
sFiles = bst_process('CallProcess', 'process_noisecov', ...
sFiles, [], ...
'baseline', [0, 547.9980469], ...
'target', 1, ...
'dcoffset', 1, ...
'method', 1, ... % Full noise covariance matrix
'copycond', 0, ...
'copysubj', 0);
% Process: Compute sources
sFiles = bst_process('CallProcess', 'process_inverse', ...
sFiles, [], ...
'comment', '', ...
'method', 1, ... % Minimum norm estimates (wMNE)
'wmne', struct(...
'NoiseCov', [], ...
'InverseMethod', 'wmne', ...
'ChannelTypes', {{}}, ...
'SNR', 3, ...
'diagnoise', 0, ...
'SourceOrient', {{'fixed'}}, ...
'loose', 0.2, ...
'depth', 1, ...
'weightexp', 0.5, ...
'weightlimit', 10, ...
'regnoise', 1, ...
'magreg', 0.1, ...
'gradreg', 0.1, ...
'eegreg', 0.1, ...
'ecogreg', 0.1, ...
'seegreg', 0.1, ...
'fMRI', [], ...
'fMRIthresh', [], ...
'fMRIoff', 0.1, ...
'pca', 1), ...
'sensortypes', 'MEG, MEG MAG, MEG GRAD, EEG', ...
'output', 1); % Kernel only: shared
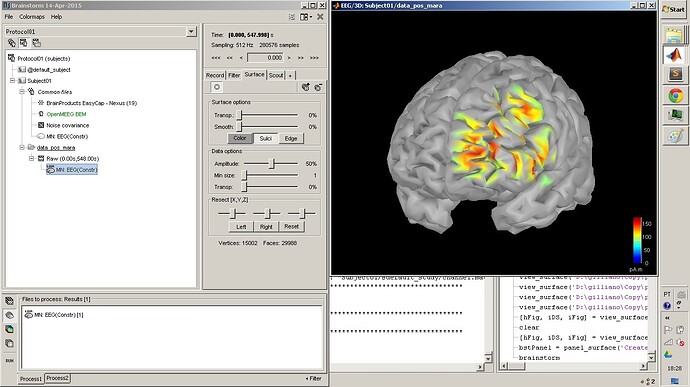
% View on the cortex surface
fn='Subject01/@default_study/results_wMNE_EEG_KERNEL_150417_1733.mat';
hFig1 = script_view_sources(fn, 'cortex');
% Set current time to 46ms
panel_time('SetCurrentTime', 0.046);
% Set surface threshold to 75% of the maximal value
iSurf = 1;
thresh = .75;
panel_surface('SetDataThreshold', hFig1, iSurf, thresh);
% Set surface smoothing
panel_surface('SetSurfaceSmooth', hFig1, iSurf, .4);
% Show sulci
panel_surface('SetShowSulci', hFig1, iSurf, 1);
% Save and display report
ReportFile = bst_report('Save', sFiles);
bst_report('Open', ReportFile);
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
When I try to run, I get this error message:
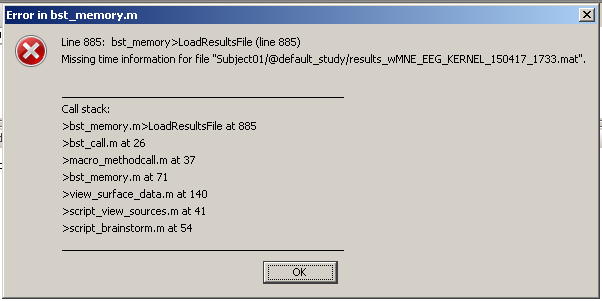
What I am doing wrong?