Dear Brainstorm-ers,
We have vitamin E pills marking the positions of 4 EEG sensors.
We do not have head shape points.
I extract the coordinates of the vitamin E pills from the T1 image (taken during the concurrent fMRI-EEG session).
This is actually a different T1 image from that provided to Freesurfer for head model construction (we align the vitamin E scans to this image).
I can load these coordinates into Brainstorm as headpoints.
However, when I try and manually align the EEG sensors (gray dots) to these headpoints (green dots), both the EEG sensors and headpoints move together, making it impossible to use the green dots as alignment markers for the gray dots.
What is the recommended approach to use these vitamin e coordinates to align the EEG and MRI?
Thank you.
-Tom.
EDIT: Is there some way I can specify the coordinates of the known sensors (those with vitamin pills attached) and have the coordinates of the other sensors updated to reflect these?
Dear Brainstorm-ers,
We have vitamin E pills marking the positions of 4 EEG sensors.
We do not have head shape points.
I extract the coordinates of the vitamin E pills from the T1 image (taken during the concurrent fMRI-EEG session).
This is actually a different T1 image from that provided to Freesurfer for head model construction.
How do I load these coordinates into Brainstorm?
What is the recommended approach to use these coordinates to align the EEG and MRI?
Thank you.
-Tom.
The only option that Brainstorm offers to register based on a few points identified both in the MRI and the EEG is using the anatomical landmarks NAS/LPA/RPA:
https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Automatic_registration
If you could see the positions of the vitamin E pills on the MRI, and if you had one pill on the forehead, one on the left and one on the wright, you could try to use the marked electrodes as the NAS/LPA/RPA landmarks. But since you use a different MRI, you can't use the standard procedure described in the tutorials.
In order to do this, you could try the following:
- Import only the T1 MRI from the FreeSurfer output
- Importing the MRI with the pills, register with SPM + reslice
- Find the coordinates in the of the 3 pills of interest
- Import the full FreeSurfer folder, use the pill coordinates as NAS/LPA/RPA
- Edit the file with the positions of the electrodes: duplicate the electrodes that are marked with Vit.E, and label the duplicates "NAS", "LPA" and "RPA".
- When importing the coordinates of the electrodes (with the menu "Add EEG positions"), the electrodes positions should be automatically converted to the SCS coordinate system: https://neuroimage.usc.edu/brainstorm/CoordinateSystems
This seems very complicated for EEG, which would not benefit much from a millimeter precision for the location of the electrodes (since you can't expect a source localization accuracy better than 1-2cm).
Here is another possible solution:
- Import the full FreeSurfer segmentation
- Import the MRI with the pills (SPM registration + reslice)
- Create a head surface using the vit E MRI -you can try tweaking a bit the parameters to see them better: https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#The_head_surface_looks_bad
- Make it the default head surface (double-click on it)
- Align manually the electrodes using this head: Show the labels of the electrodes, so that you can align the electrodes you know correspond to a vit E pill with the artifacts on the head surface
- Save the co-registration
- Make the original head surface the default again
- Edit the electrodes positions again, and project them on the head surface (the FreeSurfer one).
Does this help?
Thank you for your detailed and helpful response Francois.
I am a little stuck on one of the points.
For your first approach, you say:
- Edit the file with the positions of the electrodes: duplicate the electrodes that are marked with Vit.E, and label the duplicates "NAS", "LPA" and "RPA".
I have edited the channel file in the brainstorm_db to this end. So "Edit channel file" menu in Brainstorm now reflects this information.
Maybe you mean editing the raw data file. In our case this is a Brainvision file (.vhdr extension) with electrode coordinates (but they are all on a single plane, one of the coordinates is always 1). If I edit this file, there are no longer 64 channels but 67 and Brainstorm throws an error on import of the time series.
I am a little unclear on the goal and outcome of the approach.
I can use the provided fiducial markers (location of pills) to perform a manual alignment. It would be great if I could get the 3 electrodes (with pills attached) to default to the correct positions on the head surface (i.e. the 3 fiducials marked with the pill coordinates).
I meant: editing the file in which you have the real 3D coordinates of all the electrodes, and that you add to the channel file with the menu "Add EEG positions":
https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy#Prepare_the_channel_file
If you don't have such a file, or if you only have the coordinates of your vitamin E pills, I don't understand what you are expecting to do with only flat coordinates.
Post screen captures illustrating the data you have, that might help me understand what you are trying to do, in order to propose adequate solutions.
Thanks Francoise.
We have no channel file or head points.
We have the full 3D coordinates of 3 of the EEG sensors relative to the head surface.
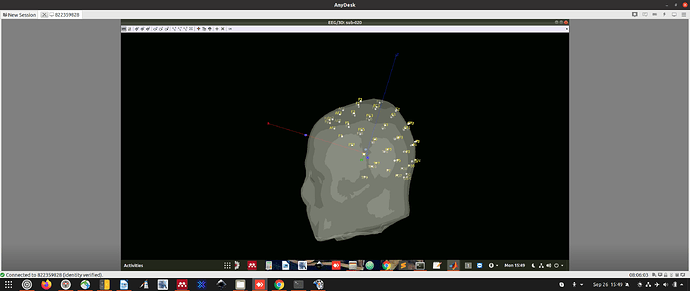
We can import the anatomy with the NAS, LPA, and RPA with the coordinates of the 3 sensors we know. This provides blue dots. We can use these blue dots to manually align the sensors (white dots) to the head surface.
Unfortunately, this is extremely time intensive (we have >100 images to align in this way) and its not easy to do. There are 9 degrees of freedom to consider. You get one sensor aligned and the others move out of position.
We were hoping there was some way to streamline the approach.
For example, if we could get the 3 sensors for which we already know the position (FPz, T7, and T8), to default to the blue dots. This would align the remaining sensors too (approximately) and provide a good starting point for manual adjustment.
Say we start with default cap electrode positions (easy cap 64). Provide the coordinates of the 3 sensors that we know and extrapolate the positions of the other electrodes (on a sphere or some such). Then load that file via menu "Add EEG positions".
What do you think?
Hopefully that helps clarify things a bit.
Let me know if you need more information.
I'm not sure what you can expect from only 3 points. Brainstorm does not offer any algorithm to reconstruct the EEG electrodes positions following the head shape, or to adjust the registration based on random sensor points.
You can use the template positions in the MNI ICBM152 space: add the EEG coordinates with the menu "Add EEG positions". I've just added a new option to adjust the MNI positions to the subject space using the linear or non-linear MNI transformation (update Brainstorm to get this new feature):
Add EEG position: Convert MNI to subject space · brainstorm-tools/brainstorm3@18e298a · GitHub
Once you have default 3D positions available in the channel file, you can write a script to compute the transformation you want to apply to these default positions (based on the electrodes for which you have the position). You can then apply this transformation to the channel file using function channel_apply_transf.m.
This would require good skills in Matlab programming, Brainstorm scripting, and 3D geometry computation. Unfortunately, this is too far from the current Brainstorm features to help you much with this project.
For loading/saving files and understanding the file structures, see the Scripting tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/Scripting
For computing the transformation based on the two sets of three points, see the code of the functions cs_compute.m and cs_convert.m.
Explanations for all the coordinate systems are available here:
https://neuroimage.usc.edu/brainstorm/CoordinateSystems
Thank you again Francoise for you replies.
To any interested reader.
We performed alignment with vitamin e pill markers (blue dots) and electrodes in one subject.
We then copied the channel file to the other subjects and then manually refined one subject at a time.