I'm using Matlab 2024a and Brainstorm 2024 (07/15) and I'm getting a strange behavior when removing ICA components. It used to be the case that when removing a component, it's time series would be replaced by a flat line and the other component time series would remain the same. Now, it seems that when removing a component, all of the components, including the one being removed, are recomputed. I should note that I'm using 28 EEG channels and 3 EOG channels. Brainstorms complains a little bit about mixing channel types, but that shouldn't be related to this behavior I think.
Thanks in advance!
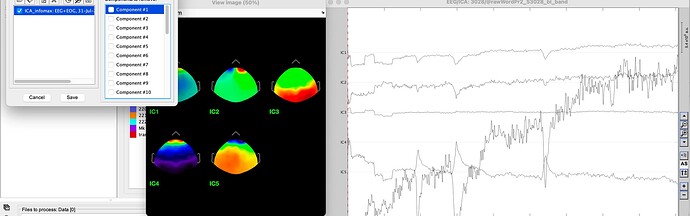
Before removing components
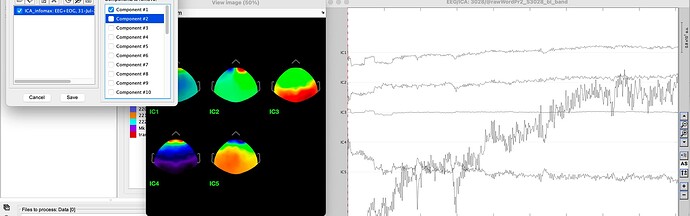
After removing component 1
Be sure that the AutoScale button ([AS]) in the IC time series figure is deactivated before removing the components.
Yes, behavior is the same with autoscale off.
Here is the expected behavior with Matlab 2019 and Brainstorm 2024 (March)
All components
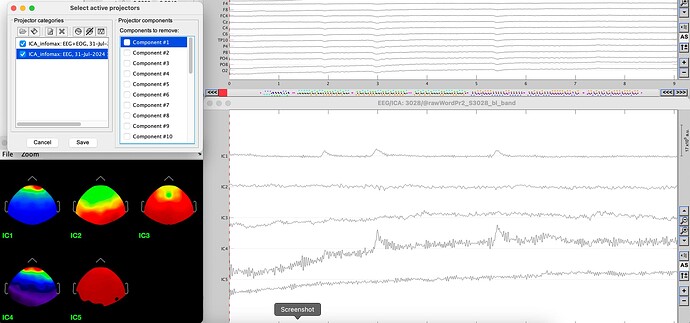
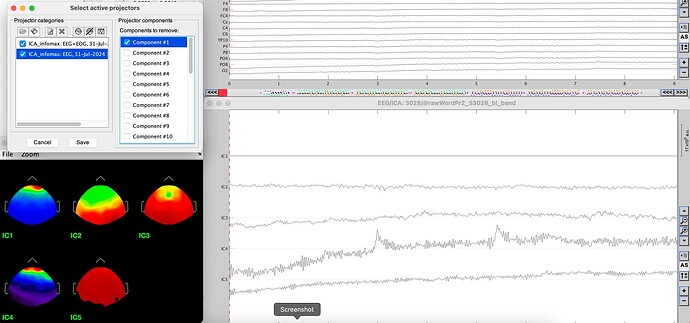
Remove Component 1
Okay, I just tried again using the same data set from my initial post and it seems it IS related to the mixing of channel types. When I use only EEG channels, I get the expected behavior. This seems unusual because most ICAs used with EEG for artifact correction should include EOG channels.
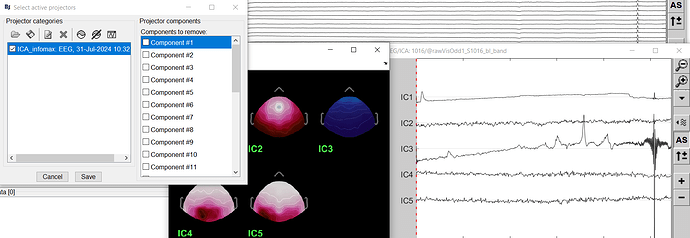
No components removed (ICA with EEG channels only)
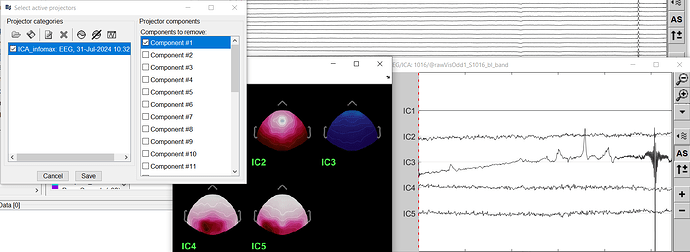
Remove component 1 (ICA with EEG channels only)
@pdkieffaber, thank you for verifying for not-mixed modalities.
We will check soon this behaviour on our side.
@Raymundo.Cassani I discovered today that this bug only happens when the infomax (EEGlab) ICA algorithm! I can use the EOG channels with the Picard algorithm and everything works as it should. I also discovered by changing my three EOG channels to EEG channels that do not have locations, that the Infomax (but not Picard) algorithm does very strange things. When I used infomax this way, the topographical maps were nonsensical and clicking to remove components did not have any affect on the data but instead seemed to alter the component time courses again. Very bizarre.
@Raymundo.Cassani, Today I discovered that the most bizarre behaviors may have been caused by the fact that I had an average reference projector based only on my EEG channels, then was trying to do the ICA with all channels.
@Raymundo.Cassani , sorry for all the messages, but as I've been working with this data, I realized that the problem is having an average reference projector on the continuous data before using the Infomax ICA. I'm not sure why it is only Infomax that is affected, but even when I have an average reference projector on continuous data where ALL channels are labeled as EEG, the function returns complex values for the component loadings and so won't display topographies, and also exhibits the bizarre behavior when trying to remove components.
@Raymundo.Cassani , Last post....I promise. It is definitely something about using an average reference projector on continuous data, then trying to use the Infomax ICA. The ICA output is nonsensical. Removing the average reference projector fixes everything.
Hi @pdkieffaber,
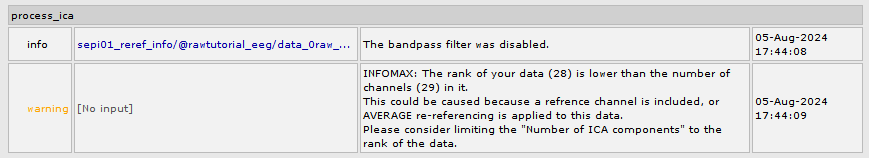
It seems the issue you are experiencing is related to the rank reduction that happens when re-referencing to the average. In the code Infomax (runica.m) does not address for it, as in EEGLab there is a popup window that tells about the issue.
Similar to Picard, now the execution of Infomax on data with a rank lesser than number of channels prints the following warning in the Matlab command window and in the report viewer. Commit 772599f
WARNING: INFOMAX: The rank of your data (28) is lower than the number of channels (29) in it.
This could be caused because a refrence channel is included, or AVERAGE re-referencing is applied to this data.
Please consider limiting the "Number of ICA components" to the rank of the data.