Hello Francois,
I'm trying to import the epoched data into Brainstorm using your alternatives, but so far without luck.
I have an SPM file with the final analyses, and I also exported it as plain .mat (which is not supported by the automatic import), EDF and EEGLAB with fieldtrip function ft_write_data and fieldtrip2eeglab (before I have to convert to fieldtrip bu this is easy).
In the EDF case I have no epochs, even though I specified in the header
hdr.nTrials = nTrials;
But Brainstrom gives me "this file does not contains any epoch"
Do you happen to know some toolbox to export EDF that is compatible with the definition of epochs that Brainstorm expects?
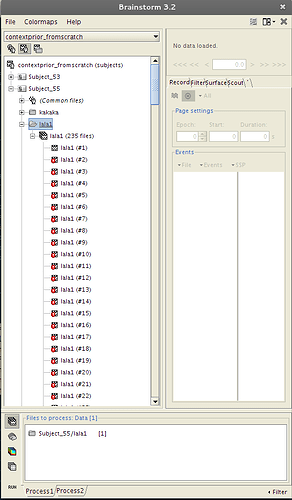
And with EEGLAB, I get these red exclamation marks in my trials (sorry for the silly test names)
and I can't select "Average Files" in the pipeline (like the trials doesn't exist)
I do manage to import the EDF file with the "Time" Version of import instead of "Existing epochs", and use the option to cut every X seconds, but the trials ends up with different times (from 0 to 1.2s, from 1.2 to 2.4s, etc...) and for instance I'm unable to calculate the noise covariance.
If I can fix that perhaps this would solve the problem, although I would feel more confortable importing with the "Existing epochs" version...
I know I could pre-process my data from the beginning with Brainstorm, but since I already have a very stable pipeline outside, with fine tunned high-pass filters and ICA artifcat removal for instance, I would prefer to just import the cleaned epochs if possible..
Also because I have and unusual way of defining conditions, which gives quite a lot of possible contrasts...
And it must be through scripts.. (not this manual one, since I need to provide the Study ID which I hope to be created during the process)
Thank you very much in advance,
Leonardo Barbosa