Dear all,
I am a professor actually in France and I am in a big neuroscience project with 3 doctoral students. We downloaded the BRAINSTORM platform to work with our simulated data (from Hodgkin-Huxley model). I am trying to import the time series dipole brain sources from matlab (3 time series .mat) to the brainstorm platform. We simulated a dipole source using the Hodgkin-Huxley 2 compartment model and it gave us 3 lines /columns of transmembrane current (Na+, K+ and Leaking). So, I need to send it to a brainstorm platform to use it within some head model (BEM, FEM or 4headmodel) to simulate the EEG signals, but after 3 weeks trying I decided to contact you to help me step-by-step. Best regards.
Hi @gzsl, you can as you may have seen in the Simulation tutorial
https://neuroimage.usc.edu/brainstorm/Tutorials/Simulations#Simulate_MEG.2FEEG_from_synthetic_dipoles
it is possible to use the process Simulate > Simulate generic signal to generate the signals for the dipole.
Since you want to use your custom signals, you could:
-
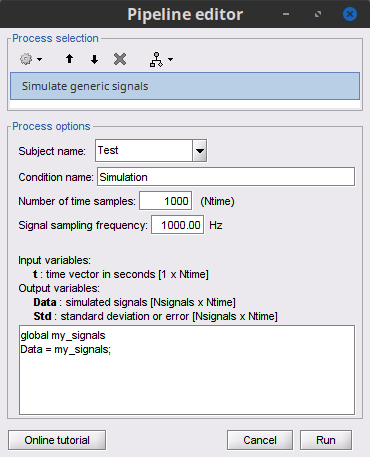
Define your signals as a global variable, e.g.,
my_signals, in the Matlab IDE:global my_signals my_signals = randn(3, 1000); -
Then use the this code in the Simulate generic signal process . Paying attention to the field Number of time samples as this needs to match the number of samples in your global variable
Alternatively, if you data is generated with a function already in the Matlab path, you can call that function directly from the Simulate generic signal process:
Data = randn(3, 1000);