Hello,
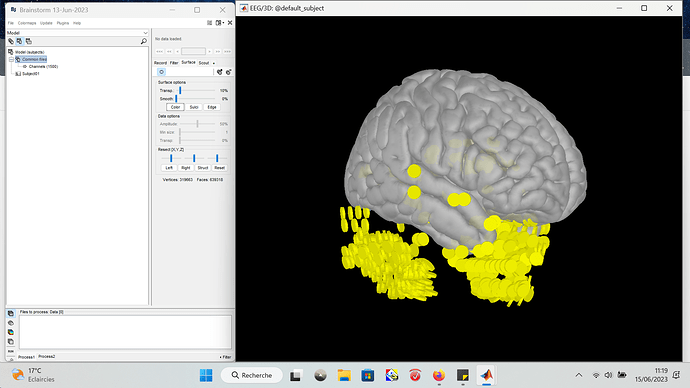
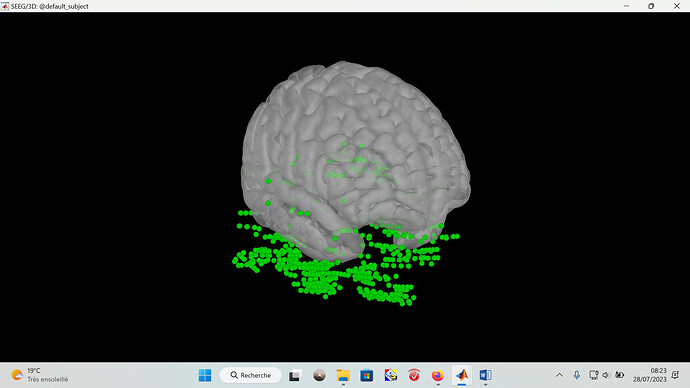
I'm trying to import my SEEG electrode MNI coordinates into an ICBM152 2023b model but it gives me this (see picture)? I do it from the EEG:ASCII XYZ_MNI,Name (,) because I only have a .txt or .csv file available... I don't understand what I'm doing wrong...
Thank you
Hi Florian,
Do you happen to have more information regarding the electrode positions?
- How were they acquired?
- What coordinate system was used?
- Where were they placed? (To verify the imported positions)
Best,
Raymundo
1 Like
Hi @Florian
Can you try another head template (one of the previous ICBM152)?
Do you have the names of the electrodes, and what will be the expected positions?
Best,
Hello @Raymundo and @tmedani
thank you for your help!! Some attempts of response. I am not in charge of analyzing the position of the electrodes. But I know that :
- it was made with Freesurfer
- in an MNI reference
- the electrodes are scattered all over the brain as in the picture
- the problem is the same with a other head template
Have a good day
@Florian, can you share your channel file?
Or at least copy-paste one line from it here in the forum?
Hello Raymundo,
thank you for your answer. Here, my data.
F
XYZ.xlsx (52.5 KB)
Thank for sharing the file!
The data in it, indeed is in MNI space coordinates, and can be imported in Brainstorm.
However, some changes are needed:
-
The file is saved as a native Excel file xlsx, it must be changed to either a comma-separated-value (csv) or tab-separated-values (tsv). This can be done from Excel. Both csv and tsv formats are text files that can be read with simple text editors such as notepad.
-
Each row should have 4 columns, Name, X, Y and Z
Find attached the same channel file in csv format, with arbitrary names.
XYZ.csv (40.7 KB)
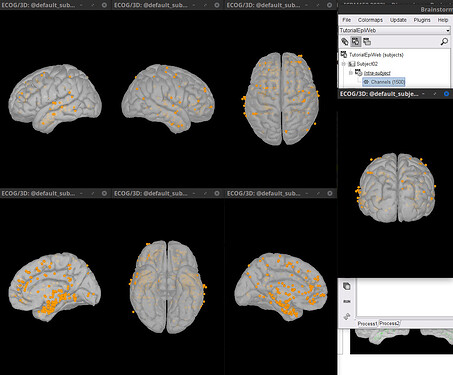
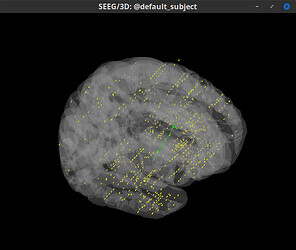
This is the imported result.
Thank you @Raymundo.Cassani
Sorry but from what import? Thank you
F
You need to select EEG: ASCII: NAME,XYZ_MNI(*.*) as Files of Type (Type de fichier)
 Because it is a text file (ASCII), the columns are Name, X, Y and Z, and it is given in MNI space
Because it is a text file (ASCII), the columns are Name, X, Y and Z, and it is given in MNI space
Hello @Raymundo.Cassani
I had used this import but I still have the same problem as at the start... which I don't understand... Thank you
That is strange 
What steps are you performing?
These are the steps I am taking:
-
Create a new Protocol
-
In Anatomy View ( ), create a new Subject using Default anatomy (ICBM152 2023b)
), create a new Subject using Default anatomy (ICBM152 2023b)
-
In Functional view ( ), right-click on Subject > Import channel file.
), right-click on Subject > Import channel file.
Select file and EEG: ASCII: NAME,XYZ_MNI(*.*) type
-
Right-click the channel file () > Edit channel file change the Type for all the electrodes to SEEG (Ctrl-A to select all, then right-click > Set channel Type)
Close and save.
-
Right-click the channel file > Display sensors > SEEG (Cortex)
1 Like
Hello @Raymundo.Cassani
Thanks, it works. I don't know where I was wrong, but it's a mistake I won't make again.
Have a good day
Florian
2 Likes