Hello I have the same problem.
I have the latest version of BrainStorm. i am trying to export my recordings from spike2 .smrx (64 channels) into .mat and i dont know how to import in BrainStorm. i always get an error saying my data is no valid.
Is it possible to import directly from spike2 .smrx file, i have seen that BrainStorm accepts .smr files, how ever this format is quite old, since several years the format is .smrx.
Many thanks in advance
Jesus
I will check if there is a free library available to read the .smrx files, and try to integrate it into Brainstorm.
Could you please share a short example file? (as small as possible)
Hello, I am Postdoc in Heidelberg University and i use Spike2 version 9 with 128 channels, and the output formaat is .smrx. I have seen that BrainStorm allow you to import files from Spike2 but only the old format .smr for 32bits. it would be great to be able to use this amazing tool directly with .smrx files.
Is there any plan to update BrainStorm to work with .smrx files?
Thank you very much for your attention and consideration.
Best Regards
Jesús
Sorry I moved around your posts before seeing that you had already started a new topic...
Please send me a small example file and all the documentation you have about the file format, and I'll see what I can do about it. Hopefully we'll be able to add a native support for the .smrx files to Brainstorm without too much trouble.
Thank you very much for you fast response, and thank you for such amazing program is Brainstorm.
i definetively can share a file in .smrx with 64 channel recordings, but i dont know how, even a file of 0.5 seconds, (2.5mb) is rejected by the system, how can i send you.
Best
Jesús
the information i have is basically the Manual from Spike version 9. also in the CED Website they provide a program to modify .smrx files in matlab, is called "MatLab/son" and y think they also have the version for python.
"The MATLAB/SON library interface is a set of files for MATLAB (Windows version) that lets users open, create and edit SON files (.smr and .smrx) in MATLAB using a script. I
This interface can also be used by any language that can link to DLLs exporting C-compatible functions" CED website.
Thank you very much
Jesus
Please upload the file somewhere (google drive, dropbox...) and post the download link here.
Thanks
My files are usually big files, so i a exported a tiny portion into .smrx. i hope is working
many thanks
Thank you for the example file.
The old SON-2.32 library we were using to read the old .smr was not capable of reading the .smrx file, therefore I had to interface the new library MATCED available for download on the CED website:
http://ced.co.uk/upgrades/spike2matson
I pushed the modifications to the Brainstorm repository:
https://github.com/brainstorm-tools/brainstorm3/commit/b18a1bf642bf01b0c6745a30a8fc788bebb89375
Please update Brainstorm and try again.
Select the file format "EEG: CED Spike2 new 64bit" file format.
This is experimental, I only had your short example file to test the reader, so there might be some bugs left in the code. Be careful with the units, and the timing of the signals and the markers. Let me know if you observe anything unexpected.
Hi Francois.
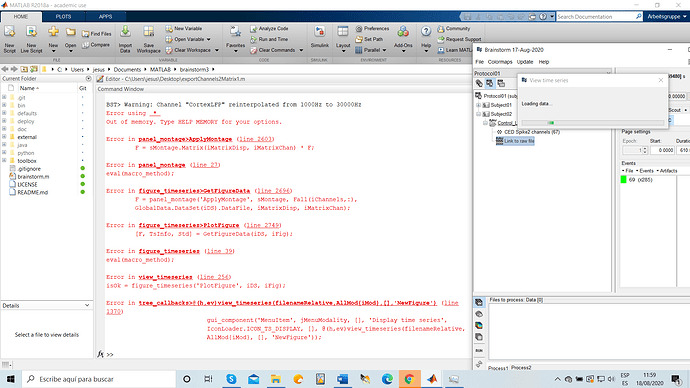
Thank you very much for the update, the example i sent you is working perfect, how ever when importing bigger files into brainstorm, i cannot open or load the "link to raw data"
actually the file i sent you was an exported .smrx file into .smrx file, in order to just to get a tiny portion of the original file. i wonder if this could be part of the problem.
below you can find full .smrx file without modifications.
https://drive.google.com/file/d/1Hsp4xpIMlfFSct0DU9bhqReKNtrUVZNK/view?usp=sharing
Indeed, you won't be able to use the menu "Import MEG/EEG" with large files.
You should use the menu "Review raw file" instead.
https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Review_vs_Import
If you are not familiar with these concepts, I recommend you start by reading all the introduction tutorials (Section "Get started", at least until #19), using the example dataset.
Some of it will not be relevant for you, but it would take you through all the basic training for using the software.
https://neuroimage.usc.edu/brainstorm/Tutorials
Thanks, for the links, i will read the tutorial thoroughly.
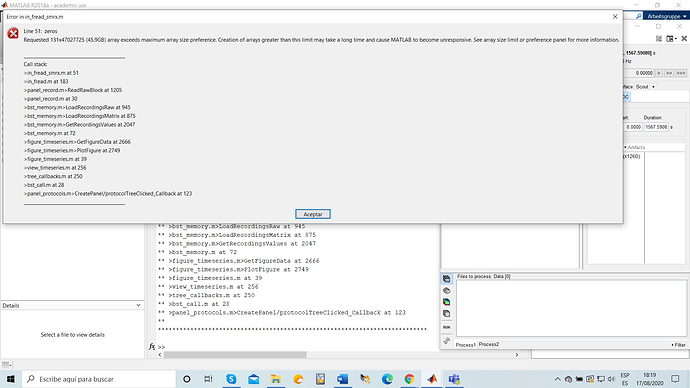
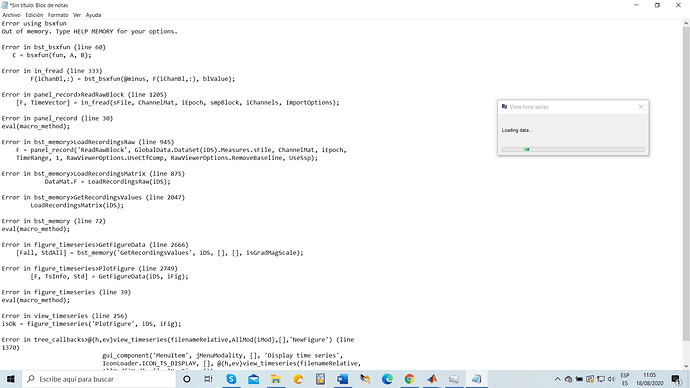
However i am all the time using the option "Review Raw file" the first thing i would like to do is to check that the file is correctly loaded, 1st option i did is to directly double click in "Link to Raw file", and i get an the error i mentioned in my previous comment. 2 option i did and then following the tutorial, right click on "Link to raw file"-->EEG--> display time series. and i get the following error.
or this error
Loading the entire file for display or for importing to the database has the same effect: you don't have enough memory to have all the data fit in your memory (45Gb), therefore you get an out-of-memory error.
When opening the recordings (double-click on the "Link to raw file"), set a shorter duration in the page settings (tab Record), for instance 30s. Then scroll through your file by pages of 30s (the tutorials will teach you how to do this), this would load only the required portions of the files at once.
Maybe your file was much smaller than 45Gb and you wonder why does Matlab want to create such a gigantic matrix in memory. Two reasons:
- the data in your file is maybe stored in int16 (16bit integers), but it's being converted to floating point double precision (64 bits) for processing and display => Size in memory multiplied by 4.
- if you have signals with different sampling rates in the same file (eg. 60 channels at 1000Hz and 1 channel at 20000Hz), loading data in Brainstorm upsamples all the signals to highest sampling rate (1kHz=>20kHz = 20x more data for 60 channels) so that the data can be represented in one single flat matrix [Nchannels x Ntime]
The Brainstorm viewer for EEG/MEG is beeing adapted to electrophysiology data, but was not initially designed for it. So we probably still have many things that are not handled as well as in your Spike2 viewer. We'll try to improve this in the future.
Thank you very much for you response, i did what you suggested and seems to work quite well using shorter times for visualization.
Another problem i am having is to import events from already "Kilosorted" data. i am using Kilosort 2 and i have many sorted neurons already that i would like to include in Brainstorm. i tried to create the structure "events.mat" is recommended in the tutorial, and it seems brainstorm read it correctly (I use : events--> import from file) however it takes very long time in importing this spikes in the database, actually it crashes very often i couldnt perform any analysis yet.
am i doing something wrong, how is should include this spikes into brainstorm for electrophysiology analysis?
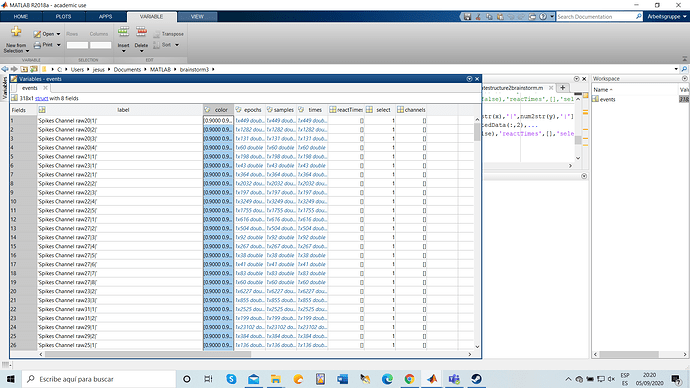
attached you will find and screenshot of the struct.mat i created following the instructions from the tutorial, and how brainstorm looks like after reading the file.
thank you very much for your help
Jesus
Could you please share your events.mat? (upload it somewhere and post the download link here)
I will try to understand what is taking so long and optimize it.
Here i uploaded the events.mat
Thank you very much
Jesus
Thanks.
I can see two problems in your file:
- The field "samples" has been removed early 2019, as it was an unnecessary duplicate of the "times" field, but source of many mismatches. I edited the screen capture in the tutorial so that it does not appear anymore. Refer to this tutorial for the definition of the events structure: https://neuroimage.usc.edu/brainstorm/Tutorials/EventMarkers#On_the_hard_drive
- The times are probably wrong in your files. Each even occurrence should align with a data sample. The multiplication
times*sfreq should lead to a list of unique integers, which is not the case in your file. To enforce aligning the times on existing samples: times=round(times*sfreq)/sfreq
Despite these two problems, I have no problem loading your event file in a similar file (30kHz, a few minutes of recordings) with the menu File > Add events from file. It takes maybe one or two second to import the file, then one or two extra seconds to display the file...
PS: I tested with Matlab 2018a on Win10 as well.
Can't you see any other warning in the Matlab command window?