Hi, brainstormers,
I am trying to load the HCP preprocessed MEG data into Brainstorm using a script because I want to load the data for all 89 subjects all at once. I used to import these files one by one through the GUI but now I would like to automate this process and transfer most of my work with the data to the cluster.
Here is the script that I generated from the process "run" box:
% Script generated by Brainstorm (12-Sep-2022)
% Input files
sFiles = [];
SubjectNames = {...
'162935'};
RawFiles = {...
'E:\files\data\162935\162935_MEG_Restin_preproc\162935\MEG\Restin\rmegpreproc\162935_MEG_3-Restin_rmegpreproc.mat'};
% Start a new report
bst_report('Start', sFiles);
% Process: Create link to raw file
sFiles = bst_process('CallProcess', 'process_import_data_raw', sFiles, [], ...
'subjectname', SubjectNames{1}, ...
'datafile', {RawFiles{1}, 'FT-TIMELOCK'}, ...
'channelreplace', 1, ...
'channelalign', 1, ...
'evtmode', 'value');
% Save and display report
ReportFile = bst_report('Save', sFiles);
bst_report('Open', ReportFile);
% bst_report('Export', ReportFile, ExportDir);
% bst_report('Email', ReportFile, username, to, subject, isFullReport);
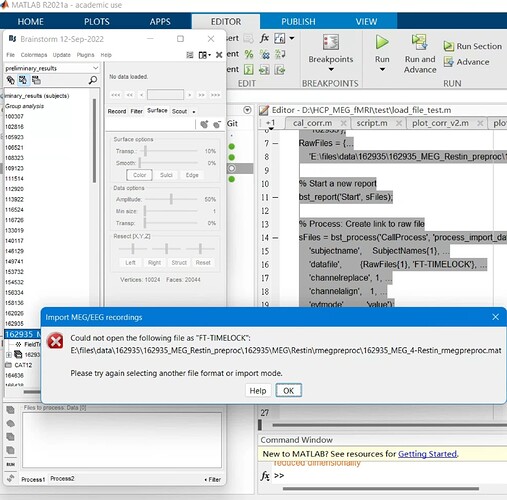
However, I am getting an error message when loading this script:
It seems like I should put in a different value in the 'datafile' parameter for the bst_process() function. Do you know what I should put in there for this type of data? I believe the HCP preprocessed data are in fieldtrip structure.
Also, is there a place where I can look for a more detailed description of each of the parameters for the process functions?
Thanks,
Andy