Hello,
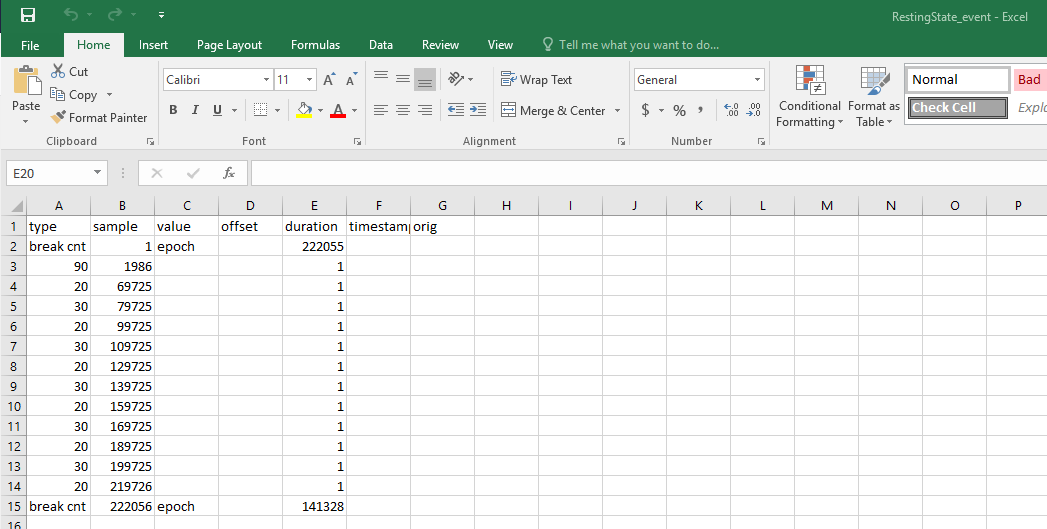
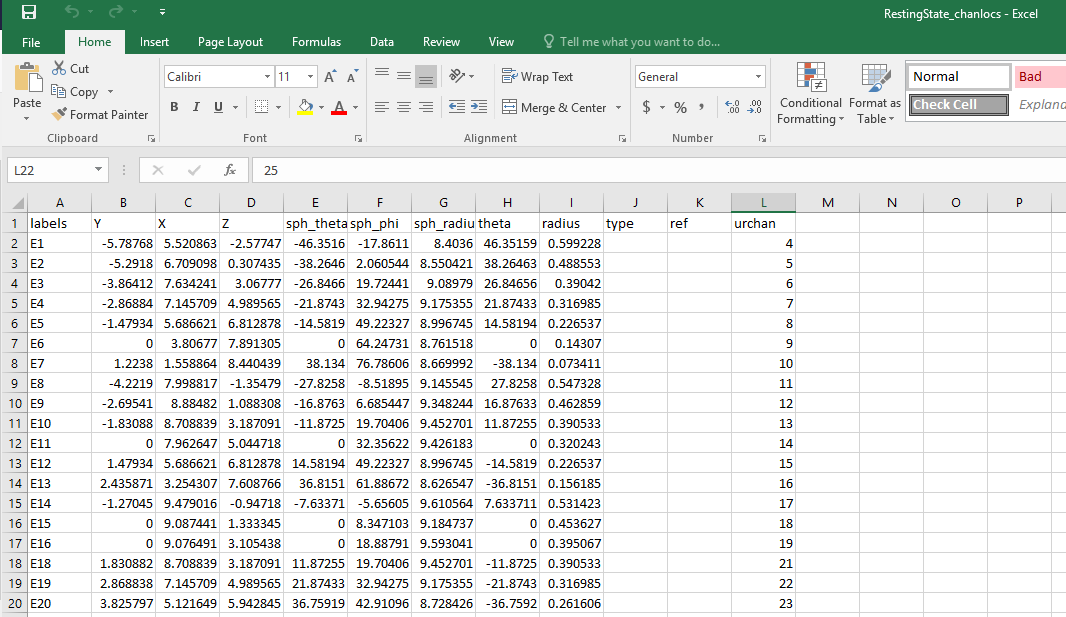
I was able to read in a .cvs signal file, but have trouble importing channel and event files, which are both in the .cvs format. I think I may need to rearrange the data structure to make the files compatible with BT. I attach two screen shots below. The data were collected by using EGI system, and sampled at 500 Hz. FYI.
Event file
I also went to File > Add events from file in the Events menu, but don't know which file type I need to select.
Channel file
What have you used to create this csv file?
From the NetStation, you can export the .mff files to the EGI .raw format or to EDF+ format. These would be better choices.
I still didn't get all the answers I needed to fix correctly the .mff reader, but I will try to push a temporary patch later today so you can keep on working.
Hello Francois,
I am currently working on secondary data analysis, and the data came from Child Mind Institute. They provide preprocessed data in two formats: mat and csv. While I was waiting for your response, I was trying to read in the .csv data. I was able to import .csv signal files, but no success with event and channel files. I also tried the .mat files, but got an error message.
BTW. the .mff file problem that I posted the other day is related to a different dataset.
What generated these .mat and .csv files?
If they were generated with custom scripts, not following any major file format, you may have to reformat these files manually to something that Braintorm can read.
Can't you ask for getting a copy of these files in a standard file format? (anything that Brainstorm can read: https://neuroimage.usc.edu/brainstorm/Introduction#Supported_file_formats
Or directly getting the original recordings and redo the pre-propressing in Brainstorm?
Hello Francois,
Both .mat and .csv files were generated and preprocessed with EGI Netstation, not custom scripts. Unfortunately, these files were downloaded from the Healthy Brain Network website, so I can't change output file formats. I tried preprocessing the original recordings in BT, but it took a long time and required many manual operations. Since I need to analyze additional 200 plus files, it would be time-efficient if I can import preprocessed data and simply conduct data analysis in BT.
I add a link to both data sets: https://www.dropbox.com/sh/yhwc4kzxyzeksrt/AACTm964cnKqCA1IEdBooWeda?dl=0
If you can help me how to import either data file, that would be great! Thanks.
Sungjin
Thank you for the file examples.
The .mat files actually contain EEGLAB structures, but not with the proper EEGLAB file names and structures. However, with very little reformatting, you can convert them to EEGLAB .set files that you can then import into Brainstorm.
a = load('WISC_ProcSpeed.mat');
EEG = a.result;
save('WISC_ProcSpeed.set', 'EEG');
Then when linking this file to the Brainstorm database with the menu "Review raw file", select the file format "EEG: EEGLAB (*.set)"
I modified a bit the EEGLAB reader in Brainstorm so these structures can be imported.
Update Brainstorm before you try this.