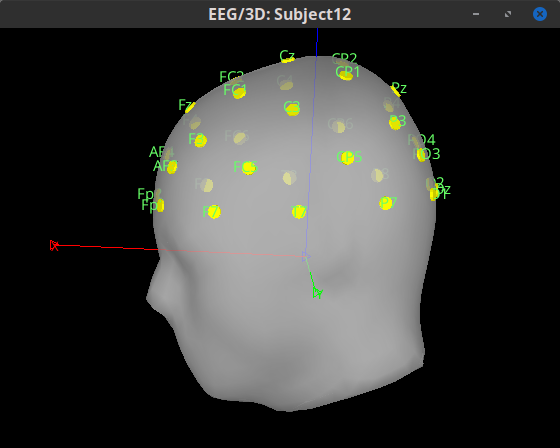
Hi!
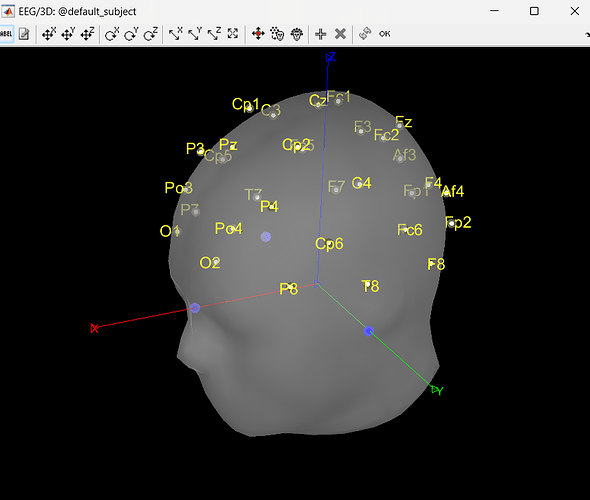
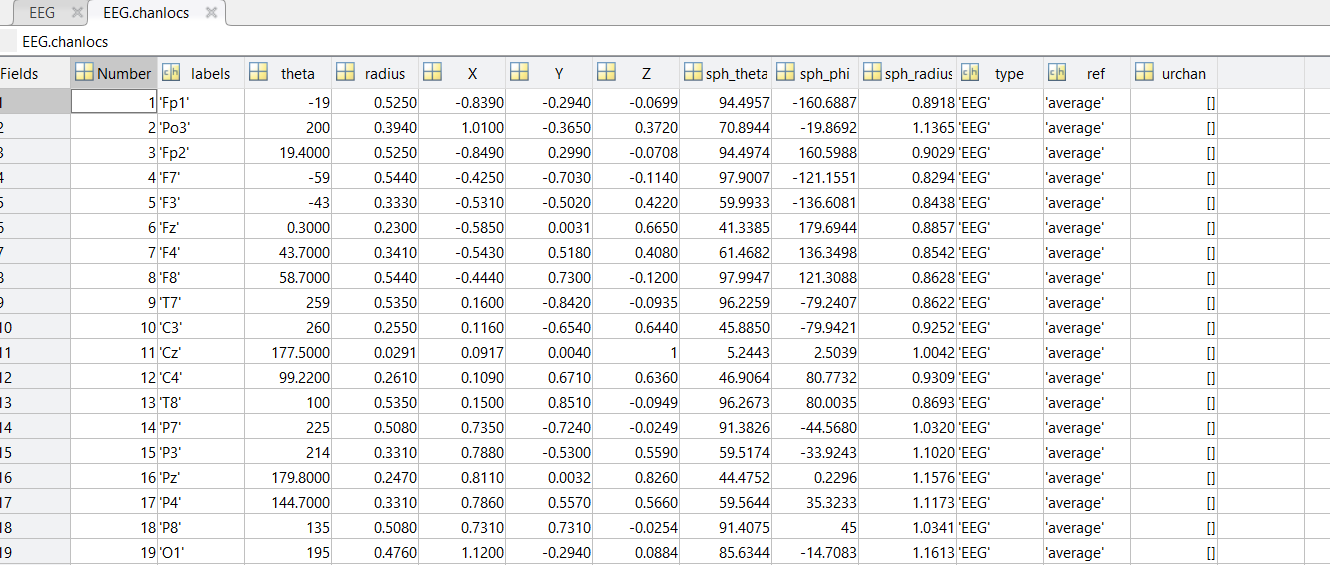
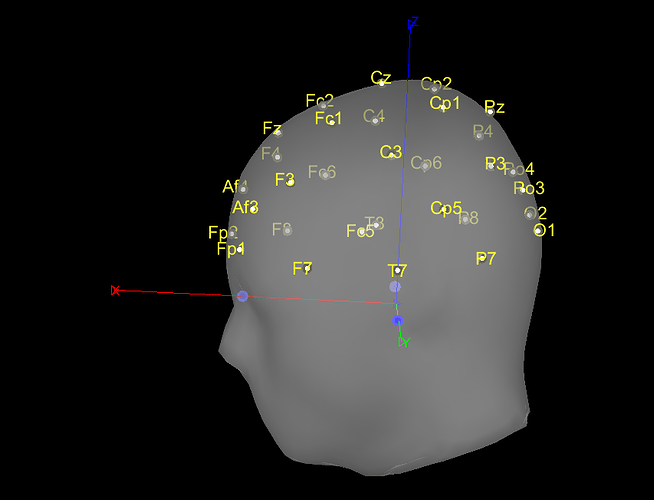
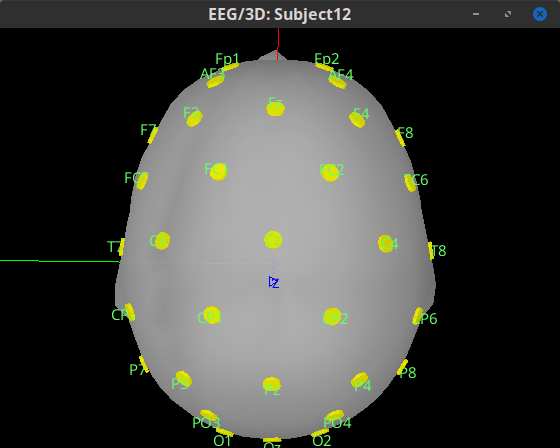
I am kinda new with Brainstorm. I currently have some pre-processed EEGs in EEGLAB format (.set). I can easily import them in Brainstorm, but, during the MRI co-registration, I noticed that they are rotated of a certain angle from the expected position (more specifically, they are rotate of about 180° or -90° among the Z axis from the desired position). Is there a way in Brainstorm, using the GUI, to correctly rotate the sensors?
I tried even extracting the information I needed using the following code:
chanlocs = readtable('Micromed31.ced', 'Filetype', 'text');
chanlocs.Properties.VariableNames = {'Number', 'labels', 'theta', ['' ...
'radius'], 'X', 'Y', 'Z', 'sph_theta', 'sph_phi', 'sph_radius', 'type'};
Positions = [chanlocs.Y chanlocs.X chanlocs.Z];
Labels = upper(chanlocs.labels);
num_chans = length(Labels);
% "Create" .elc file. Base structure taken from the epilepsy tutorial channel position file
fileID = fopen('Micromed_Brainstorm.elc', 'w');
fprintf(fileID, '#\n');
fprintf(fileID, ['# electrodes labels should be upper case by definition to' ...
' allow a proper matching of electrodes and channel labels.\n']);
fprintf(fileID, '# File generated by electrodes export feature.\n')
fprintf(fileID, '#\n');
fprintf(fileID, '# %d electrodes\n', num_chans);
fprintf(fileID, 'NumberPositions= \t%d\n', num_chans);
fprintf(fileID, 'UnitPosition \tmm\n');
fprintf(fileID, 'HSPTransformed false\n');
fprintf(fileID, 'Positions\n');
for i = 1:num_chans
fprintf(fileID, '%s:\t%.1f\t%.1f\t%.1f\n',Labels{i}, Positions(i,1), Positions(i,2), Positions(i,3));
end
fprintf(fileID, 'Labels\n');
fprintf(fileID, '%s\t', Labels{:});
fprintf(fileID,'\n');
fclose(fileID);
% Test with a struct
chanlocs_bsm = struct();
chanlocs_bsm.Label = Labels;
chanlocs_bsm.X = Positions(:,1);
chanlocs_bsm.Y = Positions(:,2);
chanlocs_bsm.Z = Positions(:,3);
% chanlocs_bsm.Type = repmat('EEG', num_chans,1);
save('Micromed31_brainstorm.mat','chanlocs_bsm');
% Test with a table
chanlocs_table = table(Positions(:,1), Positions(:,2), Positions(:,3), [1:num_chans]', Labels);
chanlocs_table.Properties.VariableNames = {'X', 'Y', 'Z', 'indice', 'name'};
a = table2struct(chanlocs_table);
save('Micromed31_brainstorm.mat','a')
I tried importing the Micromed31_Brainstorm.elc file using Adding EEG position -> Import from file (after importing the .set eeg file) in order to overwrite channels positions, with the FileType EEG:ASCII XYZ_Name or ASCII XYZ_MNI,NAME, but the warning "Warning: No channel information was read from the file." appears. I tried importing the struct or the table from Matlab workspace, but the error "Invalid structure for file type channel" appears.
Can anyone help me solve this problem?
P.S. The channel file from the epilepsy tutorial and the elc file I created are identical, if open with text editor, except from the channels and channel positions, obv