Hi there!
I'd like to compose a script that runs a whole-cortex (no scouts) Hilbert transform on source-localized epochs (already pre-processed) for all subjects. This would be like script #2
I'm having some issues with developing the script. I'm uncertain of what steps to have included in the Pipeline Editor when I wish to generate the .m script. It seems that the best way to generate the .m script would be with all steps including data importation (see image):
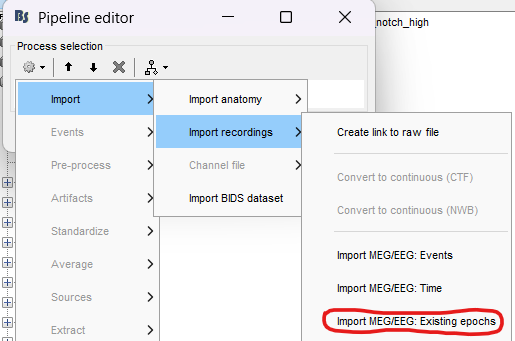
But I can't seem to import my pre-processed epochs this way since I get an error saying ***"could not open the following file as ***
"FT-TIMELOCK": \path\to\epoch001.mat
Please try again selecting another file format or import mode"
If, instead, I generate the .m script after already carrying one subjects epoch folder to the process box and setting up the Hilbert Transform for this one folder, then the script calls the subjects files as such:
% Script generated by Brainstorm (06-Apr-2023)
% Input files
sFiles = {...
- 'link|JM001/sub-001-PR90/results_PNAI_MEG_KERNEL_230310_1531.mat|JM001/sub-001-PR90/data_R1_trial001.mat', ...*
- 'link|JM001/sub-001-PR90/results_PNAI_MEG_KERNEL_230310_1531.mat|JM001/sub-001-PR90/data_R1_trial002.mat', ...*
- 'link|JM001/sub-001-PR90/results_PNAI_MEG_KERNEL_230310_1531.mat|JM001/sub-001-PR90/data_R1_trialN.mat', ...*
Is it best to continue along this latter route? I have to doubts. First, how to address the fact that all subjects have a unique amount of epochs within their epoch folder. Second, how to ensure that the bad epochs (which are still actually located in subjects respective folder) are not called within the script.
It would be absolutely amazing if I could get this script to work so as to avoid the highly time consuming alternative.
I hope this question is thorough enough and I'm happy to provide any other information that would make answering this query increasingly possible.
Warm regards,
Austin
Hi @coopapap,
Where are these epochs coming from?
The error indicates that the epochs were tried to be imported as FieldTrip files.
If you want to compute the Hilbert transform for each source file in each subject, you can drag-and-drop all the Folders from all the Subjects at the same time. One Hilbert transform file ( ) will be created for each source file. Before computing the Hilbert transform for many source files check this section in the TimeFrequency tutorial, as this process can easily generate gigantic files:
) will be created for each source file. Before computing the Hilbert transform for many source files check this section in the TimeFrequency tutorial, as this process can easily generate gigantic files:
https://neuroimage.usc.edu/brainstorm/Tutorials/TimeFrequency?highlight=(hilbert)|(storage)#Full_cortical_maps
Best,
Raymundo
Hi Raymundo!
Thank you for this.
I am importing epochs from within each subjects Brainstorm folder. They are evidently in .mat format, and the only option I see that uploads .mat epochs is that of Fieldtrip:
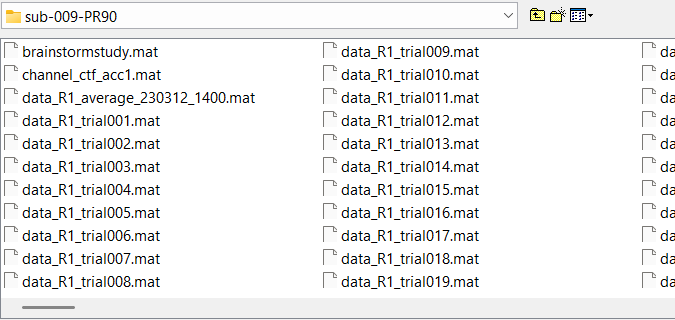
I want to perform a Hilbert transform on each individual subjects epoch folder (not the average epoch,, but the folder with each epoch in it). I haven't found a means of doing this through the pipeline editor (I have only found a means of computing a group-Hilbert transform through this method.
I think a script is the best way but I still don't know the best way to proceed.
I suppose the first question is, is there a means of importing epochs of this kind through the pipeline editor, as I have tried to do?
If they are in Brainstorm folder, they is not need to import them again.
What's the reason you want to (re-)import them?
This is possible as described in the previous post.
- Drag-and-drop the folders containing the epochs to the Process1 tab
- Select the Sources button(
 ), the legend
), the legend Files to process will indicate how many source (result) files are found in all the folders
- Select the process Frequency > Hilbert transform
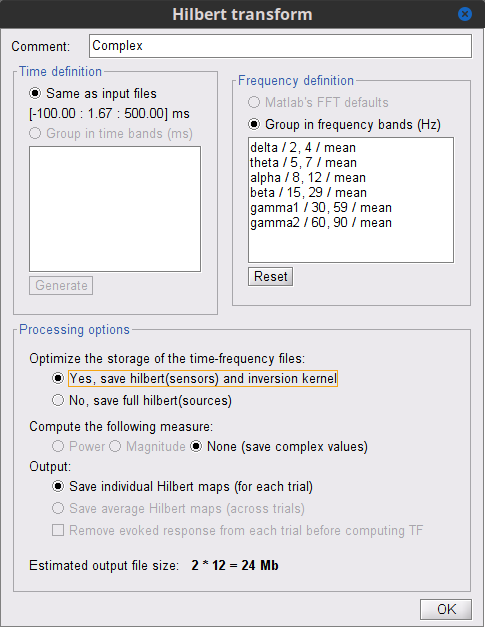
- Before running the process, click on the Edit.. button of the Hilbert transform options to:
a. Define Time and Frequency options
b. Optimize storage (Select Yes if the option is available)
c. Select the desired measure (The only option is None if Optimize storage option is Yes)
d. Verify that the Output will be the Individual Hilber maps (for each trial) (The only option is Save individual if Optimize storage option is Yes)
Hi Raymundo!
Thanks for this!
I thought that I needed too import the data through the pipeline editor so as to automate the process through the "Generate .m script" option.
And regarding the method you propose for the Hilbert Transform, I have tried this way but since I'm uploading the epoch folders that have each epoch (not the average of all epochs per subject) then it will compute a Hilbert for each epoch (so approximately 30 Hilbert Transforms/subject). This is too much.
Again, thank you so much for your help Raymundo!
Cheers,
Austin