Hello @Raymundo.Cassani
Thank you for your answer, the link provided was helpful to get things started.
Each subject (n = 27) went through 8 acquisition runs therefore the same analysis pipeline needs to be looped over the 8 runs over the 27 subjects. Hence, Loop over acquisition runs is better suited for what I am trying to do.
What do you think?
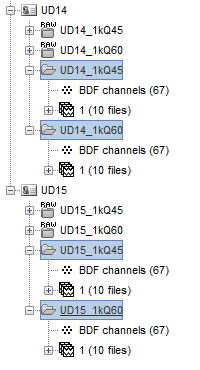
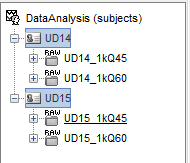
Here is a simplified snapshot of what I am trying to explain. In this case, I am only showing 2 subjects each with 2 acquisition runs. What needs to happen is that the same analysis pipeline needs to loop over the 2 runs AND over the 2 subjects.
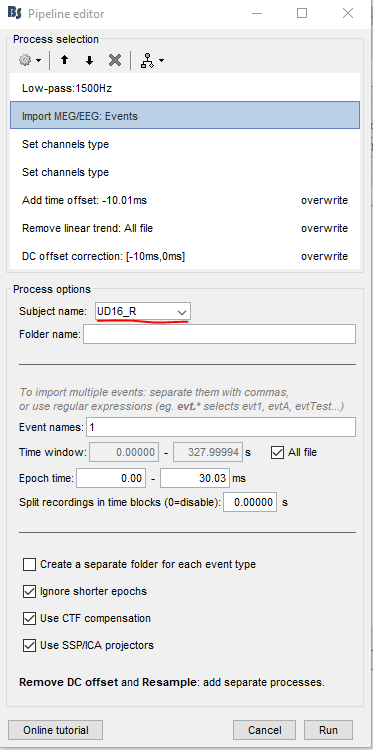
The process that is causing difficulty from the pipeline is the "Import MEG/EEG: Events" process. Therefore, to go through some trial and error for this specific process, I made the following script using the 2 subjects that I showed above. The script was made using the step-by-step instructions from the Loop over acquisition runs tutorial section. The script is as follows. There are two nested "for loops" to loop over both RUNS and SUBJECTS.
% Script generated by Brainstorm (17-Jul-2021)
% Input files
% Added the other subject names and all the runs for all the subjects
% as per the tutorial instructions
SubjectNames = {...
'UD14',...
'UD15'};
sFiles = {...
{'/.../@rawUD14_1kQ60/data_0raw_UD14_1kQ60.mat', ...
'/.../@rawUD14_1kQ45/data_0raw_UD14_1kQ45.mat'}, ...
{'/.../@rawUD15_1kQ60/data_0raw_UD15_1kQ60.mat', ...
'/.../@rawUD15_1kQ45/data_0raw_UD15_1kQ45.mat'}};
% Loop on subjects
% Added as per the tutorial instructions
for iSubject = 1:length(SubjectNames)
% Loop on runs for each subject
% Added as per the tutorial instructions
for iRun = 1:length(sFiles{iSubject})
% Start a new report
bst_report('Start', sFiles);
% Process: Import MEG/EEG: Events
sFiles = bst_process('CallProcess', 'process_import_data_event', sFiles, [], ...
'subjectname', SubjectNames{iSubject}, ...
'condition', '', ...
'datafile', {sFiles{iSubject}{iRun},'MAT'}, ...
'eventname', '1', ...
'timewindow', [], ...
'epochtime', [-0.2002, 0.5996], ...
'createcond', 0, ...
'ignoreshort', 1, ...
'usectfcomp', 1, ...
'usessp', 1, ...
'freq', [], ...
'baseline', []);
% Save and display report
ReportFile = bst_report('Save', sFiles);
bst_report('Open', ReportFile);
% bst_report('Export', ReportFile, ExportDir);
end
end
Unfortunately, I am facing some issues with this script with numerous attempts not going anywhere other than receiving error messages like the one below.
>> script_new
Operator '==' is not supported for operands of type 'cell'.
Error in file_win2unix (line 31)
if (FileName(1) == '/') && ~file_exist(FileName)
Error in bst_process>GetInputStruct (line 1464)
FileNames = cellfun(@file_win2unix, FileNames, 'UniformOutput', 0);
Error in bst_process>CallProcess (line 2185)
sInputs = GetInputStruct(sInputs);
Error in bst_process (line 37)
eval(macro_method);
Error in script_new (line 25)
sFiles = bst_process('CallProcess', 'process_import_data_event', sFiles, [], ...
Hence, my conclusion is that just following the Loop over acquisition runs tutorial section is not enough to make this work (unless I misunderstood the tutorial material).
What do you think I should try now?