Hello,
I am currently trying to replicate the following article in Brainstorm: https://www.sciencedirect.com/science/article/pii/S0165027024001389
Nicely, the author put the data in BIDS online: OSF | Localizing hidden Interictal Epileptic Discharges with simultaneous intracerebral and scalp high-density EEG recordings
I was wondering if you could help me import it in brainstorm.
1/ I noticed that there was no dataset_description.json file needed by brainstorm so I created one from an other study:
{
"Name": "",
"BIDSVersion": "1.7.0",
"DatasetType": "raw",
"License": "",
"Authors": [
"",
"",
""
],
"Acknowledgements": "",
"HowToAcknowledge": "",
"Funding": [
"",
"",
""
],
"EthicsApprovals": [""],
"ReferencesAndLinks": [
"",
"",
""
],
"DatasetDOI": "doi:"
}
2/ The EEG data are in derivative while anat is in the root folder
Brainstorm doesn't import the data from derivatives. So I copied the anat folder from the root to the derivatives
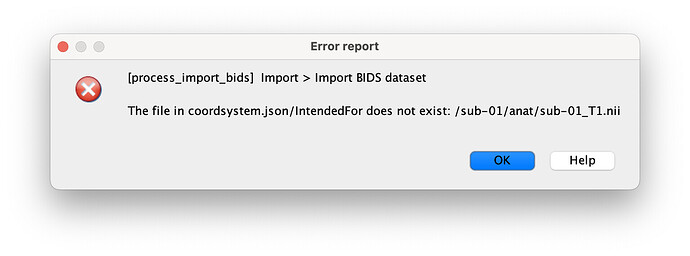
3/ Issue with the channel
I get the following error:
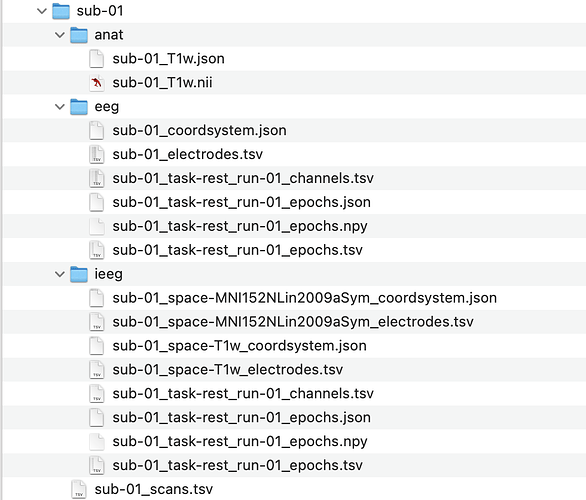
The subject folder looks like this:
So the file contains sub-01_coordsystem.json:
{
"EEGCoordinateSystem": "T1w",
"EEGCoordinateUnits": "mm",
"AnatomicalLandmarkCoordinates": {
"LPA": [
-71.0715,
5.01604,
-19.5648
],
"NAS": [
25.1881,
89.3942,
14.9043
],
"RPA": [
78.7842,
-7.95071,
-16.9084
]
},
"AnatomicalLandmarkCoordinateSystem": "T1w",
"IntendedFor": "/sub-01/anat/sub-01_T1.nii",
"AnatomicalLandmarkCoordinateUnits": "mm"
}
Edit: I tried to remove the line IntendedFor but then it doesn't import anything. or to change it to "/sub-01/anat/sub-01_T1w.nii" or sub-01_T1w.nii without success
I think we manage to replicate the article that it can be a nice tutorial for Brainstorm. I'd be happy to help writting it if you want ![]() (note: actually; it seems they are not sharing the entire SEEG recording but only the data of one contact so less interesting ... still interesting to see if we can import it :))
(note: actually; it seems they are not sharing the entire SEEG recording but only the data of one contact so less interesting ... still interesting to see if we can import it :))
Edouard