Hey Everyone,
I want to automate the manual definition of fiducial points (NAS/LPA/RPA) on the Anatomical MRI (instead of clicking on related points on the MRI).
MRI files were analyzed by FreeSurfer before uploading to Brainstrom.
We have accurate fiducial points (NAS/LPA/RPA) defined during TMS navigation by Brainsight software.
Below, the "bst_process" function uses "nas," "lpa," and 'rpa' parameters as input:
bst_process( ...
'mrifile', {AnatDir, 'FreeSurfer'}, ...
'nas', [127, 213, 139], ...
'lpa', [ 52, 113, 96], ...
'rpa', [202, 113, 91]);
Everything is OK. But, the fiducial points (NAS/LPA/RPA) defined by Brainsight provide those points locations in different spaces.
I want to transform one of the below coordinate systems to the coordinate system that is used by bst_process function. However, I couldn't find any transformation matrix or function.
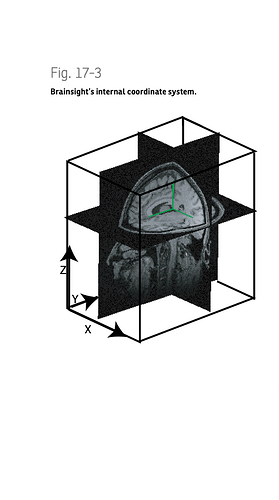
Please see below the coordinate systems' names and exported fiducial points (NAS/LPA/RPA) examples by Brainsight. All are indicated same points in different spaces.
Brainsight Location System
'Nasion' 'Session 2' 97.3729000000000 36.5679000000000 116.260500000000
'RPA' 'Session 2' 169.700900000000 140 80.3649000000000
'LPA' 'Session 2' 24 136.624600000000 96.9871000000000
Nifti Scanner
'Nasion' 'Session 2' 0.130100000000000 111.879100000000 -25.2005000000000
'RPA' 'Session 2' -72.1979000000000 8.44700000000000 -61.0961000000000
'LPA' 'Session 2' 73.5030000000000 11.8224000000000 -44.4739000000000
LOCATOR (.elp/.hsp)
//Position of fiducials X+, Y+, Y- on the subject
%F 0.105419 1.42109e-17 0
%F 0.00344251 0.0732616 0
%F -0.00344251 -0.0732616 0
BESA (.sfp)
FidNz 0.00 10.53 -0.00
FidT9 -6.84 0.00 -0.00
FidT10 7.83 0.00 -0.00
I also added not exact but approximate positions of fiducial points (NAS/LPA/RPA) as defined by manual clicking on the MRI viewer of Brainstorm (desired approximate locations on Brainstorm x,y,z):
(NAS/LPA/RPA)
NAS: MRI: 130 220 109 - World: 2.5 111.45 -32.46
RPA MRI: 202 139 95 - World: 74.5 30.45 -46.46
LPA MRI: 48 108 96 - World: -79.50 0.55 -45.46
Could you please help me transform one of the above coordinate systems to the Brainstrom coordinate system?
This issue has discussed before, but I think never solved:
Here, the defined brainsight coordinate system.
Best,
Mehmet