Hi everyone!
I would like to investigate inter-regional phase-amplitude coupling between two scouts (each composed by 200 vertex) to see how much phase of slow oscillations (8-12Hz) in one scout drive the power of faster rhythms (30-45Hz) in the other one.
After computing irtPAC, I obtained a matrix 200x13x39 that I opened in matlab: 200 are the vertex, 13 the time bins, 39 the high frequency bins.
I was wondering how could I average all the 200 vertex of the scouts (if you think it's appropriate) and how could I see the low frequency bins.
Could anyone please help me with that?
Thank you in advance!
Sarah
Hi Sarah,
Can you tell me how you did it: which process and options selected - or a screenshot of the process options? I'm not really understanding the size of the result, how you'd get 200 vertices from 200x200. I'll check what the code actually does with what you selected.
Otherwise, a simple approach is to do dimension reduction with a scout function before computing PAC.
Best,
Marc
Hi Marc,
thank you so much for your reply.
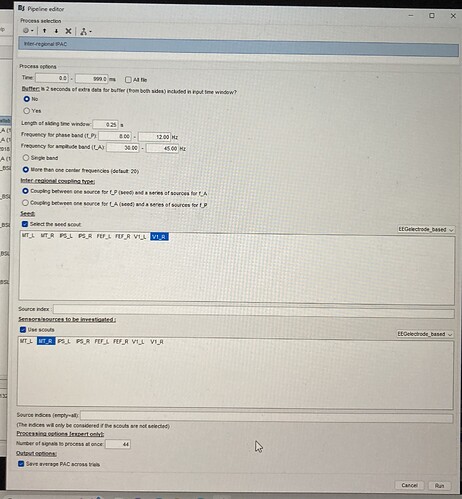
Here you can see the options that I selected, maybe I’m doing it wrong.
Hope to hear from you!
Thank you,
Sarah
Hi Sarah,
I looked through the code and now understand a bit more what it's doing. Sorry for the very limited documentation. First, the "seed scout" label is misleading because it actually only uses one source time series (one vertex) from the selected scout, the one marked as the seed for the scout (I forget if it's possible to view it through the scout panel, but it's saved in the atlas). Furthermore, it doesn't deal properly with unconstrained sources (bug) - so it would pick a somewhat random single component (among x,y,z) of a somewhat random vertex within the scout.
So in your example, one time series is used from V1_R, and tPAC is computed with all the vertices of M1_R. This explains the resulting array dimensions.
As for averaging, it's a bit complicated. The tPAC process doesn't actually save full comodulograms, but it applies a peak-finding procedure on a file-by-file basis (e.g. each trial) among the frequencies for phase (low frequencies). So each trial, and each signal/vertex is treated individually and a single different f_P is selected and saved. This is an issue that has come to our attention recently so we don't yet have a solution. (I'm also not yet clear on what it does when the peak-finding procedure fails.)
I suggest you follow this thread on Github that I just created: tPAC issues · Issue #618 · brainstorm-tools/brainstorm3 · GitHub And we can update this post if/when there's a better answer.
Let us know if you have further questions or comments in the mean time.
Marc