Hello,
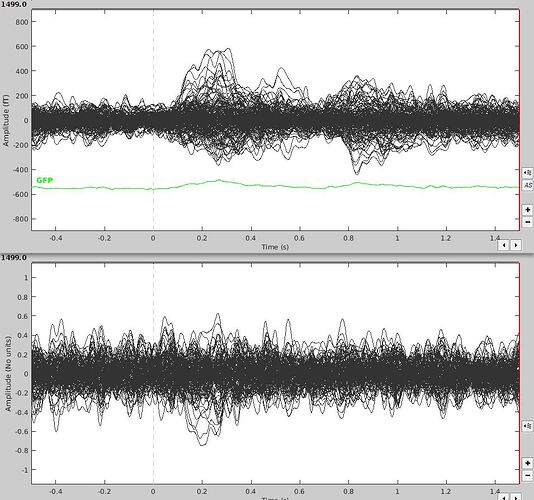
I am dealing with time locked data and have a baseline of 500 ms. When I average my trials, the time series show a relatively clear response contrasting with the baseline. However, when I compute the sources (LCMV, median eigenvalue, constrained), the baseline appears far less different than the stimulus window at scout-level. Is that something I can expect?
Here is an example with a graph showing the average time series above, and all scout means of the source of that same average (I'm not sure how comparable it is as it's more an accumulation of source means than an overall mean, but I don't know if I can display a mean whole-brain source space time series).
Doing baseline standardization helps. However I'm looking at computing the difference between stimuli, and your "Difference" tutorial suggest I add baseline z-score normalization at that point. When I do it seems only to transform it into a z-score (leaving the noisy baseline untouched). Maybe I just don't understand what it's doing?
I also have another question: is it possible to do a baseline standardization on the time series before computing the source of the signals? No source is being computed on the z-score standardized average when I attempt that. Is it doing the same thing if I do a baseline standardization at source level (so after computing sources)?
Many thanks,
Charly