Hi,
Greetings!
I am working on the meg data and I want to get the brain source data including time series (4D volume data). When I tried to export the sLORETA data to Matlab, I got the data shown in the picture below. I am wondering where I can find the time series data as I can just find the data of the grid location.
For another, I wanted to export the same sLORETA data as Nifti format, so I used "Export to SPM8(volume)" to export the data. I chose keeping time dimension (4D volume) and the time downsample factor and volume downsample factor are both 1. I don't know why it took hours to export the data. I guessed it is because the file is big (the final exported .nii data is 1,409,400,001 KB). When I opened the data in ITK-SNAP, I found the resolution of the image is about 2.5 mm, while I chose "regular grid 5mm" when I computed head model (MRI volume). I hope to know why the resolution changed when the data was exported.
Thanks a lot for the attention.
Best,
Junjie Ma
Hello,
The file you show contains an inverse operator (ImagingKernel), it has to be multiplied by some recordings to obtain the source time series. This will not be a 4D volume, but the time series corresponding to each of you dipole in your volume.
This is properly documented in the introduction tutorials:
http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Computing_sources_for_single_trials
http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#On_the_hard_drive
The source files are never full volumes in Brainstorm. The sources are estimated for a cloud of source points (it can be a regular grid, the cortex surface, or any list of 3D points), and are interpolated on the MRI for display.
A full MRI with 2x2x2 mm voxels may have more than 3,000,000 voxels, while the number of grid points used for estimating the minimum norm is in the range of 30,000.
I don’t recommend you export the full time resolution / full spatial resolution results as a volume, because it generates gigantic files (as you have already noticed), but also because it does not correspond to what you want to do in SPM.
If you export your data to .nii files, it is probably for computing statistics across subjects or across conditions. You should compute your measure of interest (eg. average a short time window) before you generate these .nii files. Otherwise, downsample heavily the time or spatial dimensions.
Francois
Hi Francois,
Thanks for the reply. I am wondering what it means that the volume does not correspond to what I want to do in SPM.
Since the SPM8 is not the proper way to export the sLORETA data, what kind of way do you recommend if I want to export the sLORETA data (4D data with time series at every voxel) and make it into a 4D numpy array for further statistically processing in python or matlab. I prefer the data without downsampling the time or spatial dimensions.
Thanks a lot for the attention.
Best,
Junjie Ma
You can export the full volumes if you don’t mind saving 50Mb of information in files that are over 1Tb.
Otherwise, simply work with the source time series (ImagingKernel * Data), or average in time, or downsample.
Hi Francois,
In your response “You can export the full volumes if you don’t mind saving 50Mb of information in files that are over 1Tb.” Do you mean using SPM8(volume) to export the 4D data like what I did before? If the answer is yes, what should I do to make the resolution of the exported data 5mm instead of 2mm? Or the 2mm resolution is the default value for the data exported by SPM8(volume) that I can not change?
Thanks for the attention.
Best,
Junjie Ma
5mm is the resolution of the source grid, it is completely unrelated with the volume on which the source values are re-interpolated after. The resolution of the output volume is the same as the one of the anatomy you defined for this subject (the file “MRI” in the anatomy part of the protocol).
To decrease the spatial resolution of the output volume, use the option “Volume downsample factor”. If you enter “2” here, it will save one voxel every two voxels. If your subject MRI volume is 2x2x2mm, the output will be 4x4x4mm.
Hi Francois,
Thanks for the reply. I kind of got the point how the SPM 8 works. As I just wanted to export the source grid data so I can have time series at regular grid points. Is there a way to export without interpolation?
Besides, as you said the source files are estimated from a cloud of source points instead of the full volumes, what is the criteria to choose the voxels used to estimate the source? And how can we forces it to use the same voxels for estimation for different subjects?
Another question is that how I can load the nifti data into Brainstorm which is exported from Brainstorm using SPM8.
Thanks for your attention.
Best,
Junjie
As I just wanted to export the source grid data so I can have time series at regular grid points. Is there a way to export without interpolation?
The ImageGridAmp (or ImagingKernel * Data) contain the time series corresponding to the grid points in GridLoc:
http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#On_the_hard_drive
what is the criteria to choose the voxels used to estimate the source?
Voxels refer to elements of a full grid, here we are using a grid of point that is not necessarily isotropic and that usually do not sample the full volume but only the brain. The various options are described here:
http://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource
And how can we forces it to use the same voxels for estimation for different subjects?
You can use load the same grid for all the subjects, but we usually recommend to use a source model that matches the anatomy of the individual subject instead. For group analysis, see the following tutorial:
http://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects
http://neuroimage.usc.edu/brainstorm/Tutorials/TutWarping
Hi Francois,
I am very grateful for your kind help. As there are two ways to get the ImageGridAmp data, I tried both of them.
Firstly, I tried to get the full source maps including ImageGridAmp in Brainstorm, and error reported: "index exceeds matrix dimensions". The details are shown in the picture below.
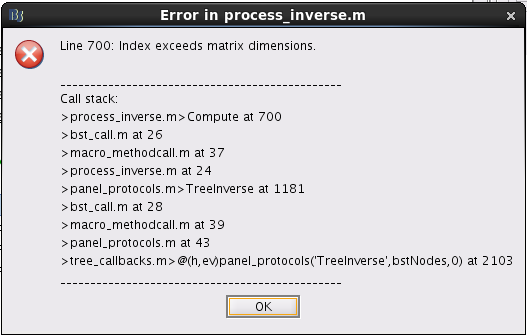
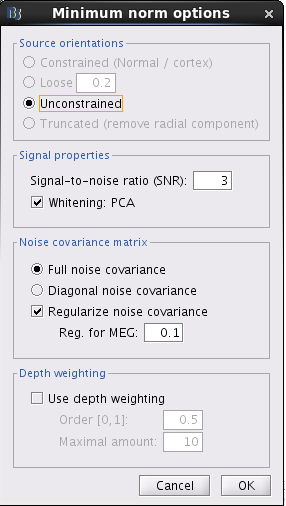
Secondly, I followed the step you put forward in another posts to calculate the ImageGridAmp using imagekernel, which is shown below.
- Right-click on the file to load > File > Copy file path to clipboard
- In the Matlab command window: ResultsFile = '....paste the file path with CTRL+V...';
- ResultsMat = in_bst_results(ResultsFile, 1);
Then Error message returned: "Cannot use this operation on source files attached to continuous RAW files."
In my case, the source data is sLORETA:MEG(Unconstr). What's wrong with my data and procedure? Just let me know if you need more details. I am so sorry for keeping bothering you and thanks very much for your time.
Best,
Junjie Ma
Indeed, the continuous input files are not designed to be the files you are directly working on.
Import a short block of recordings first (right-click > Import to database), copy-paste or recompute your head model, and finally compute and export your source maps.