Dear customers,
I'm looking for plot location of EcoG grid on the brain.
in edit channel file in the "location" I cannot understand which kind of value do I have to type (MNI, SCS, MRi?).
Alternatively, after imported my ecog raw data for each channels, can I type their location manually on the 3D cortex?
Thanks in advance,
Luca
Hi Luca,
All the coordinates saved in Brainstorm structures are in SCS coordinates, including the positions in the channel file:
http://neuroimage.usc.edu/brainstorm/CoordinateSystems
I don’t recommend you edit the positions of the electrodes in this way. You can mark them on the MRI viewer, as illustrated in this SEEG tutorial:
http://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity
There are still some issues with ECOG, but we should fix them soon and write another tutorial specific for that other type of data. Start by following this tutorial with the example dataset, then when working on your own data, immediately after linking your continuous recordings to the database, edit the channel file and set the types of the appropriate channels to “ECOG”.
Thank you Francois,
I m working on that following your sugestions!
Is there any opportunity to edit EcoG position not by MRviewer, but by means 3D render of the patient?
Luca
Yes, you can right-click on the channel file > MRI Registration > Edit… (Cortex)
But you will only see the center of the contact, not the full rendered contact.
Read the tooltips of the buttons in the toolbar for help.
Thanks again Francois.
I see the command, but is available only to add contacts, not to edit from the beginning my grid position.
To do that I have before to import channel file. May be something wrong in my procedure…
Luca
Indeed, this is only to tune existing positions.
Start by adding your various grids with the panel iEEG, in the MRI viewer, as illustrated in the tutorial.
Then use this 3D view to modify the positions.
Note that I’ve been fixing many bugs related with ECOG in the past few days, and I will do a few more edits today. Start by updating Brainstorm, and keep an eye on the updates log: http://neuroimage.usc.edu/brainstorm/Updates
Thanks Francois!
Luca
Dear Francois,
I have a question related to this issue. I'm trying to define ECoG electrodes and encountered some bugs.
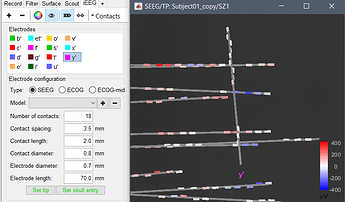
First: If I switch in the iEEG panel from sEEG to ECOG I still see all the sEEG Standard electrodes and the set skull entry option etc.If I add a new electrode under the ECOG Option I can start setting the first electrode of the Grid.
Second: In the moment I'd like to add the second point (at the second corner) I get an error (Line 2039: ./ Matrix dimensions must agree ...)
Any help is appreciated!
Best Johannes (former jpzoellner  )
)
First: If I switch in the iEEG panel from sEEG to ECOG I still see all the sEEG Standard electrodes and the set skull entry option etc.If I add a new electrode under the ECOG Option I can start setting the first electrode of the Grid.
Fixed: Bug fixes: Various GUI issues in iEEG panel for ECoG · brainstorm-tools/brainstorm3@e550924 · GitHub
Menu Update > Update Brainstorm, and try again.
Second: In the moment I'd like to add the second point (at the second corner) I get an error (Line 2039: ./ Matrix dimensions must agree ...)
This issue, I don't manage to reproduce. After updating Brainstorm, if you still get the same error, please post an exact sequence of operations that leads to this error, together with the full error message (including the stack trace).
Thanks for helping us debugging the ECOG part of this iEEG panel.
Francois
Hi Francois,
thank you for the fast bufix!
Unfortunately the second problem is not solved.
I load the MRI, set the Fiducals, load the CT and start the "Implantation". I can set an sEEG electrode without problems (see screenshot).
When I try to set the Grid I can set the first point, but in the moment I'd like to set the second point I get the error (see screenshot).
Any ideas?
best, Johannes
I still don't manage to reproduce this error on my computer. I don't understand how this error can occur...
Can you detail the exact sequence of clicks you do starting from the creation of the new grid (maybe I'm not doing it in the same order as you)?
What version of Matlab are you using? (type "ver" in the Matlab command window for full information)
I will probably need some help to debugging this. If you are not too afraid by the Matlab editor and debugger, could you try the following:
- Open function brainstorm3/toolbox/gui/panel_ieeg.m in the Matlab editor
- Place a breakpoint at line 2073 (click on the left of the line in the editor, on the dash right next to the line number)
- Start the creation of your electrode grid, do everything until the moment it is supposed to crash.
- Instead of getting the error message, the debugger will stop at your breakpoint and you can explore the contents of the variables. Please post here the output of the following commands in your Matlab command window:
sElectrodes(iElec)
w1
w2
sElectrodes(iElec).Loc
size(sElectrodes(iElec).Loc(:,1) * w1 + sElectrodes(iElec).Loc(:,2) * w2)
size(w1 + w2)
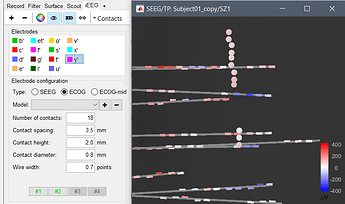
If you want to create an ECoG grid 4*8, you should enter "4 8" or "8 4" in the field "Number or contacts", instead of 32. It would let you set 4 four points: pick the middle of the contacts at 4 corners of the grid.
Sorry for the lack of online documentation at the moment, we will write a proper tutorial for ECoG at some point, when we get data we can redistribute. Having a proper test dataset would help a lot for validating the tools. Btw, if you do have a dataset that would be adapted for an online tutorial and would like to contribute to the project, you'd be more than welcome to join us, similarly to what we've done with Freiburg and Grenoble in the other clinical tutorials:
https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy
https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity
Hi Francois,
I use a matlab 2012b….
I click in the Anatomy on the CT and choose Implantation -> defining the first contact of the Grid in the MRI viewer -> click on the “+” -> adding electrode “test” -> click on ECOG -> chose a defined 8x4 Grid (now with “8 4”, 10mm spacing and a diameter of 4mm) -> click on #1 -> choose the second Grid point (below the 1) -> click on #2 -> define the 3 point, click on #3 -> chose the 4th point opposite to the 2nd and click on #4
Now I get an error “line 1890 error('No inner skull or scalp surface for this subject.'”
In the workspace I have the variables ChanLoc (32x3), SurfaceType ‘ECOG’ and sSubject
I don’t have a surface file loaded in brainstorm. Is this necessary? I just load MRI and a CT as I'm only interested in the MNI coordinates (at the moment) and perform the further processing directly in matlab.
After setting the “8 4” options properly for 32 electrodes I don’t get the error I described first.
I send you a Mail regarding the last part of the post.
Thanks and best, Johannes
Hi Johannes,
For now, the positioning and the display of the ECOG grids depends on the cortex and inner skull surface. When you create your grid by setting the 4 corners, it creates a flat grid and then projects it on the inner skull. This is something that could be changed, designing a tutorial would help for identifying and making work the most standard workflows.
For now, you have two options:
- either you set your grid as "ECOG-MID" in the iEEG tab, and this should not crash if the surfaces are missing (if it still does, this needs to be fixed),
- or you compute default surfaces for the patient: right-click on the subject T1 MRI > SPM canonical surfaces. You need to have SPM12 set up on your computer for using this functions, or you can use the compiled version of Brainstorm, which includes the SPM functions needed for this (see the section "without matlab" of the installation page: https://neuroimage.usc.edu/brainstorm/Installation)
François
Hi François,
thank you for the fast reply. I'd be happy to contribute to a tutorial! Did you get my mail?
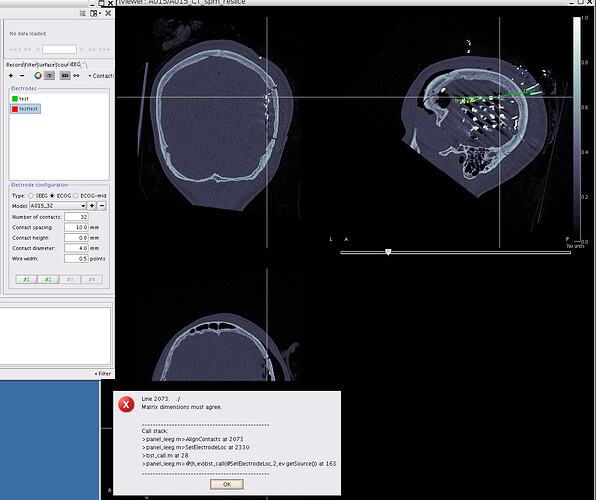
If I perform the 2nd option and compute the surface I get the following error:
** Error: Line 2174: ./
** Matrix dimensions must agree.
**
** Call stack:
** >panel_ieeg.m>AlignContacts at 2174
** >panel_ieeg.m>SetElectrodeLoc at 2330
** >bst_call.m at 28
** >panel_ieeg.m>@(h,ev)bst_call(@SetElectrodeLoc,4,ev.getSource()) at 165
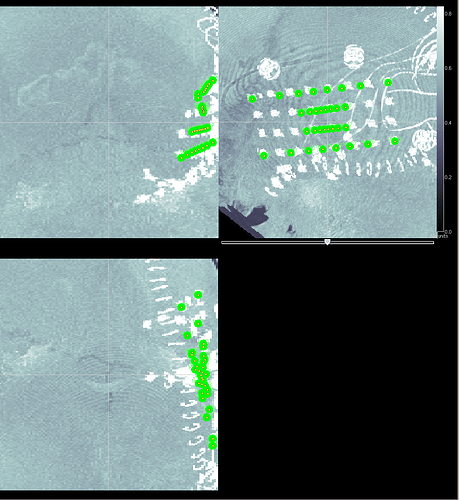
if I perform the 1. option the Grid looks like the screenshot. Maybe I didn't got the order of the corners correct. The upper left Grid point is electrode 1 (#1) the upper right electrode 8 (#3) the lower left electrode 25 (#2) and the lower right electrode 32 (#4).
(
)Best, Johannes
... additional comments:
-
If you switch from the ECOG to the ECOG-mid Option you can't select the Grid Model specified in the Type: ECOG, but if I'd like to define a Grid with the same name you are not allowed to do it.
-
If you start implanting a electrode (also sEEG) and don't finish or it fails, you can still see the "Implantation" under "sEEG (subjects)" but can't open it on the CT again, so you have to delete it and start again.
If I perform the 2nd option and compute the surface I get the following error:
I've just fixed many small things, including the possibility to place the contacts without a surface (it would be a flat grid and you'd have to move the contacts one by one though...)
Please update Brainstorm and see if it works any better.
if I perform the 1. option the Grid looks like the screenshot
You should select the grid corners clockwise or counter-clockwise. Here it looks like you selected a corner, and then its diagonal. You should try selecting the channels in this order:
#1: Contact 1
#2: Contact 8
#3: Contact 32
#4: Contact 25
If you switch from the ECOG to the ECOG-mid Option you can't select the Grid Model specified in the Type: ECOG, but if I'd like to define a Grid with the same name you are not allowed to do it.
I fixed this, now you should have the same list for ECOG and ECOG-mid.
If you start implanting a electrode (also sEEG) and don't finish or it fails, you can still see the "Implantation" under "sEEG (subjects)" but can't open it on the CT again, so you have to delete it and start again.
To edit an existing channel based on the CT: right-click on the channel file > MRI registration > Edit (MRI Viewer : CT)
Otherwise, if you do this from the anatomy, right-click on the CT > SEEG/ECOG implantation, it starts a new implantation.
Is there anything not working at this level?
Thanks for helping me debuging this!
Dear Francois,
using the ECOG-mid option it works. Thank you!
If I use the ECOG option it still crashes (even with a surface file):
** Error: Line 2317: ./
** Matrix dimensions must agree.
**
** Call stack:
** >panel_ieeg.m>AlignContacts at 2317
** >panel_ieeg.m>SetElectrodeLoc at 2489
** >bst_call.m at 28
** >panel_ieeg.m>@(h,ev)bst_call(@SetElectrodeLoc,4,ev.getSource()) at 165
best, Johannes
Indeed, I didn't test this with strips, only with grids with two dimensions...
I fixed a few more bugs: https://github.com/brainstorm-tools/brainstorm3/commit/f860f1fad38bf278a41d315de4de1b1fc7466d7b
Please update, try again, and let me know how it goes.
Thanks for helping me making this work!
It works impressively well using the SPM canonical surface estimation!
No need at all to adapt the electrodes like in the ECOG-mid option.
Thank you very much!