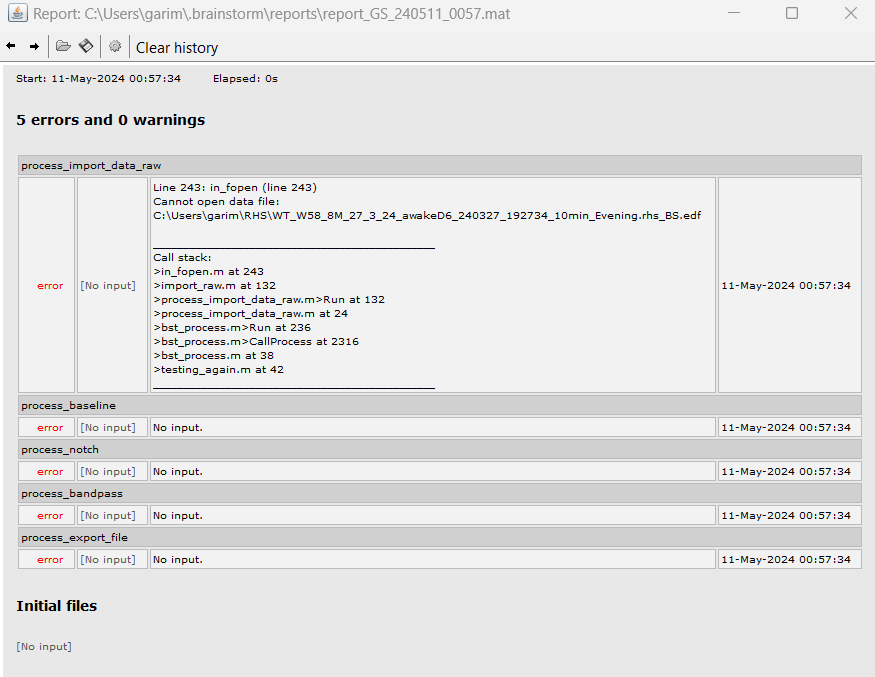
this is the code I am using:
folder = 'C:\Users\garim\RHS';
all_files = dir(fullfile(folder, '*.rhs')); % Get all .rhs files in the folder
if isempty(all_files)
disp('No .rhs files found in the folder.');
else
new_filenames = cell(length(all_files), 1); % Preallocate cell array to store new filenames
for i = 1:length(all_files)
old_filepath = fullfile(folder, all_files(i).name);
new_filepath = [old_filepath '_BS.edf']; % Append "_BS.edf" suffix to the original filename
movefile(old_filepath, new_filepath); % Rename the file
new_filenames{i} = new_filepath; % Store the new filename with full path
end
end
% Update all_files to include full paths
for i = 1:length(all_files)
all_files(i).name = new_filenames{i};
end
% Display the updated original filenames and the new filenames
disp('Updated Original Filenames with Full Paths:');
disp({all_files.name}');
disp('New Filenames with Full Paths:');
disp(new_filenames);
%RawFiles = cell(length(all_files), 1); % Preallocate cell array to store pairs of filenames
for i = 1:length(all_files)
sFiles = [];
SubjectNames = {...
'NewSubject'};
RawFiles = {all_files(i).name, new_filenames{i}};
% Start a new report
bst_report('Start', sFiles);
% Process: Create link to raw file
sFiles = bst_process('CallProcess', 'process_import_data_raw', sFiles, [], ...
'subjectname', SubjectNames{i}, ...
'datafile', {RawFiles{i}, 'EEG-INTAN'}, ...
'channelreplace', 1, ...
'channelalign', 1, ...
'evtmode', 'value');
% Process: DC offset correction: [0.000s,total duration]
sFiles = bst_process('CallProcess', 'process_baseline', sFiles, [], ...
'baseline', [0, ], ...
'sensortypes', 'MEG, EEG', ...
'method', 'bl', ... % DC offset correction: x_std = x - μ
'read_all', 0);
% Process: Notch filter: 50Hz
sFiles = bst_process('CallProcess', 'process_notch', sFiles, [], ...
'sensortypes', 'MEG, EEG', ...
'freqlist', 50, ...
'cutoffW', 1, ...
'useold', 0, ...
'read_all', 0);
% Process: Band-pass:1Hz-70Hz
sFiles = bst_process('CallProcess', 'process_bandpass', sFiles, [], ...
'sensortypes', 'MEG, EEG', ...
'highpass', 1, ...
'lowpass', 70, ...
'tranband', 0, ...
'attenuation', 'strict', ... % 60dB
'ver', '2019', ... % 2019
'mirror', 0, ...
'read_all', 0);
% Process: Export to file: Raw
sFiles = bst_process('CallProcess', 'process_export_file', sFiles, [], ...
'exportraw', {RawFiles{i}, 'EEG-EDF'});
% Save and display report
ReportFile = bst_report('Save', sFiles);
bst_report('Open', ReportFile);
% bst_report('Export', ReportFile, ExportDir);
% bst_report('Email', ReportFile, username, to, subject, isFullReport);
% Delete temporary files
% gui_brainstorm('EmptyTempFolder');
end
It doesnt solve the issue plus gives an error: