Hello Everyone, I need help in anomalizing fMRI file for my research, so that I can use it in Brainstorm. I am looking for MATLAB program.
P.s I don't have good knowledge in MATLAB programming.
Thank You, all suggestions are highly appreciated!!
Hello
Brainstorm is not a suited for processing fMRI data.
If you are looking at solutions for anonymizing MRI volumes, you can check out the following programs:
- FreeSurfer / mri_deface: https://surfer.nmr.mgh.harvard.edu/fswiki/mri_deface
- Brainstorm: Cut plane based on MNI registration (right-click on MRI in database > Deface volume): https://neuroimage.usc.edu/brainstorm/News#March_2019
- FieldTrip: Interactive selection of a volume to cut: http://www.fieldtriptoolbox.org/faq/how_can_i_anonymize_an_anatomical_mri/
- PyDeface: https://github.com/poldracklab/pydeface
- XNAT face masking: http://nrg.wustl.edu/software/face-masking/
Hi Francois,
Apologies that I was not clear in my message to you.
I have received some MRI data (T1) in DICOM file from a hospital we collaborate. The data contain the patient information (name, data of birth). I can import these volumes (DICOM files) in Brainstorm and everything works fine.
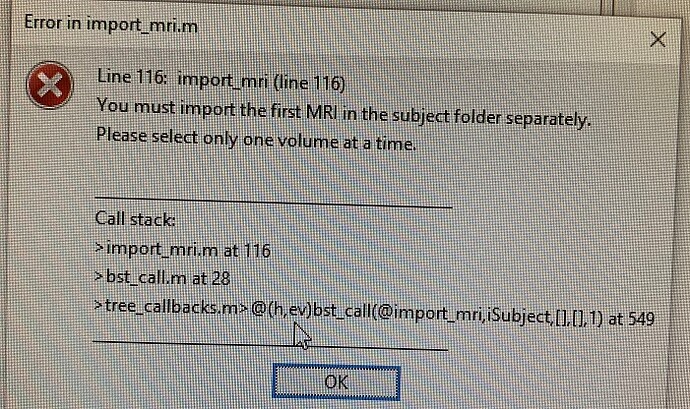
However, I would like to anonymize the files and trip the data from patient sensitive information before importing them in Brainstorm. I have used the function dicomanon to anonymize the slices. The function works fine and the outcome files are indeed anonymized. I can open the volume in MRIcro fine. However, when I try to import the anonymized (processed data) into Brainstorm, I got some dialogue box (P.s which I have attached as image file) which basically considers the MRI image folder as different volumes and second dialogue box which tells us to "select one volume at a time" .
Can you please suggests how can I fix this issue.
Thank you!!!
However, I would like to anonymize the files and trip the data from patient sensitive information before importing them in Brainstorm
To import DICOM data directly into Brainstorm, you need to select the file format "MRI: DICOM (SPM converter)". This calls directly the SPM12 DICOM converter (which must be installed on your computer and up-to-date), and imports the .nii generated by it.
Why the anonymized DICOM folder are causing SPM to produce multiple volumes instead of only one: I don't know. This would be a question to ask the SPM developers.
If you load your DICOM volumes into Brainstorm with the SPM importer, you may not need to anonymize them before. Most of the DICOM info is already stripped by SPM when converting to .nii, and then Brainstorm keeps only some parts of the .nii info. Check what is left in the MRI structure loaded in Brainstorm by right-clicking on the MRI > File > View file contents.
The volumes can be additionally defaced and anonymized in Brainstorm: right-click on the MRI > Deface volume. You brought my attention to the fact that this step was not always removing all the sensible information from the original .nii file header. I added a few lines of code to the defacing process to scrub all the possible identifying information from the data structure:
Anatomy: Defacing MRI now deletes the History and Header info · brainstorm-tools/brainstorm3@b23359f · GitHub
As a summary, you can import the original fully identified DICOM in Brainstorm, use this menu Deface volume, and finally export the anonymized volume as a .nii file (right-click > File > Export to file).
However, when I try to import the anonymized (processed data) into Brainstorm, I got some dialogue box (P.s which I have attached as image file) which basically considers the MRI image folder as different volumes and second dialogue box which tells us to "select one volume at a time" .
You should have a dialog box asking you which files you want to import. Just do what this error message suggests, select only one volume the first time. And if you need the other ones, call the DICOM converter a second time.
PS: To make screen captures on windows, you can simply press the PrintScreen key on your keyboard, or ALT+PrintScreen for capturing only the active window. Then you can paste the captured images directly in the forum text box or in any other program (CTRL+V).
Thank you for your help Francois, I was able to alter my algorithm in MATLAB and able to sort out my issues.