- How do I understand the meaning of headshape.fid.pos? Does it have anything to do with SCS coordinates?
I'm sorry, I'm not competent to tell you how FieldTrip reading functions work exactly, or what you would get from reading a FIF file with ft_read_headshape. Please refer to the FieldTrip documentation or the FieldTrip support channels (Support - FieldTrip toolbox) for more information.
When reading files in Brainstorm, everything is converted to CTF/SCS which is clearly documented in the online tutorials:
https://neuroimage.usc.edu/brainstorm/CoordinateSystems
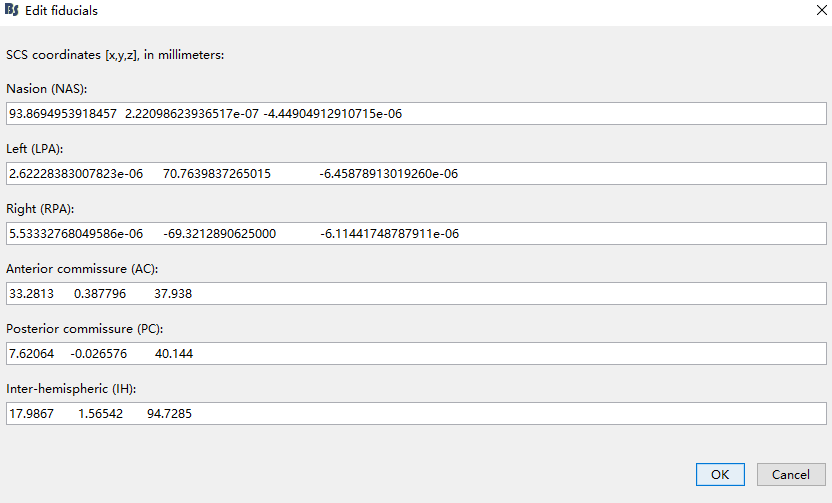
- Can I specify the fiducials position of MRI as headshape.fid.pos? Just like this,
You can, but it sets the MRI fiducials, not the MEG fiducials, so I'm not sure what you'd expect to do here. Make sure you understand clearly all the documentation about coordinate systems and MEG/MRI co-registration before trying to play randomly with the registration information. This is a critical point of the MEG processing pipeline, which is also highly prone to misunderstanding and manipulation errors.
https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Automatic_registration
Does this mean that it is not feasible to specify the financial position of MRI as headshape.fid.pos?
The fiducials coordinates you want to use (from this .fif file) are the coordinates that were digitized during the MEG acquisition. They make sense only if they are used together with the MEG sensors coordinates and all the extra digitized head points. Using them arbitrarily for any other purpose than applying a spatial transformation to the MEG sensors+head points does not make sense.
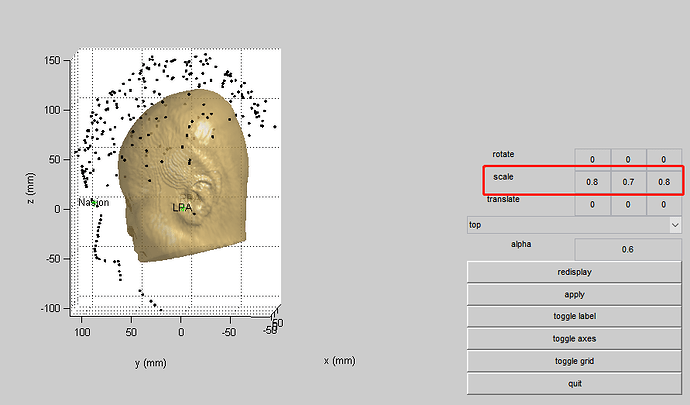
- After I reassigned the coordinates of the MRI fiducial points, I got the following registration results:
As explained, I don't think this makes much sense.
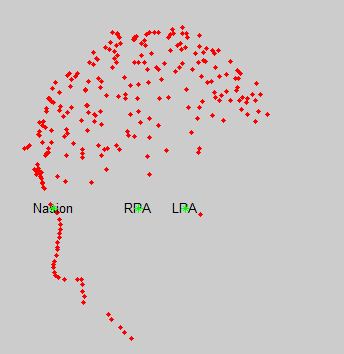
- After that, I removed points below nasion and refined using head points. I got the following registration results:
As mentioned in my previous post: if you use this automatic registration based on the head shape, all the refinements you can bring to the NAS/LPA/RPA fiducials before (your step #3) are useless. The previous MRI-MEG registration is completely replaced.
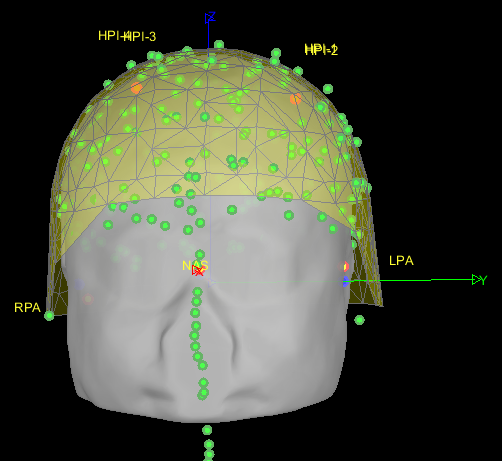
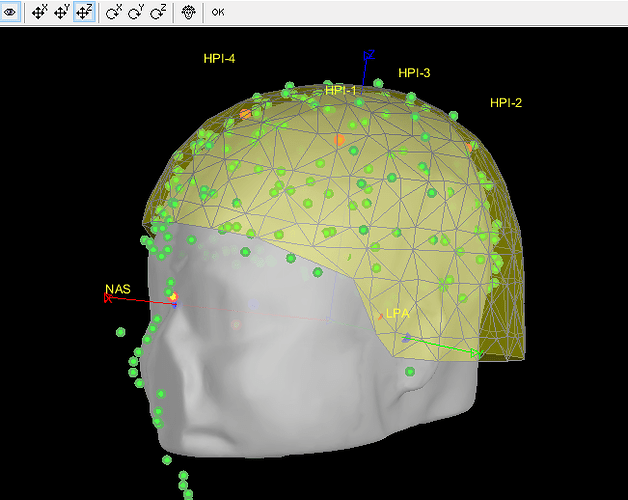
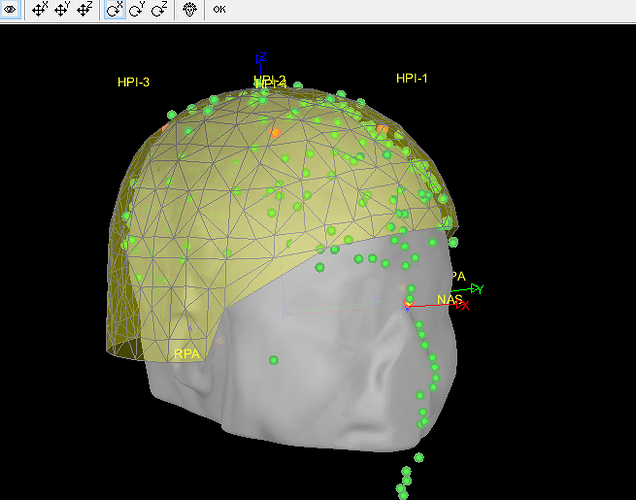
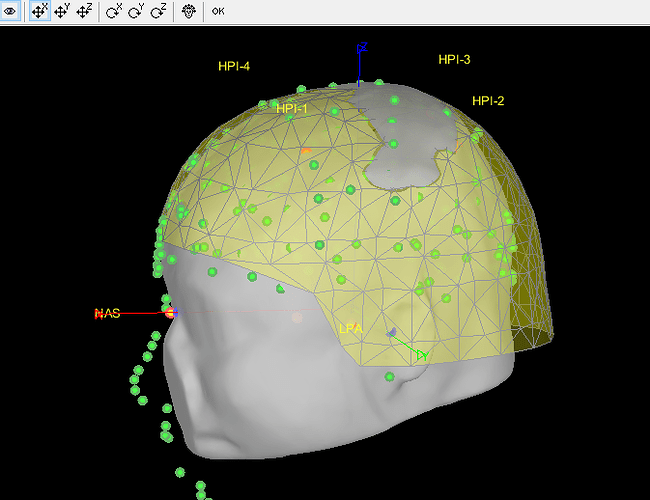
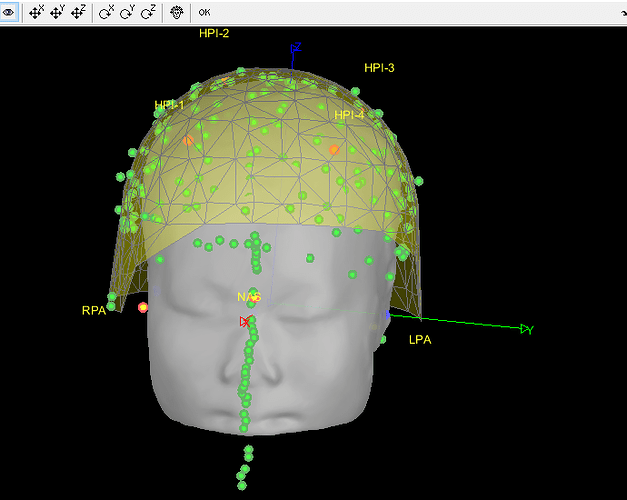
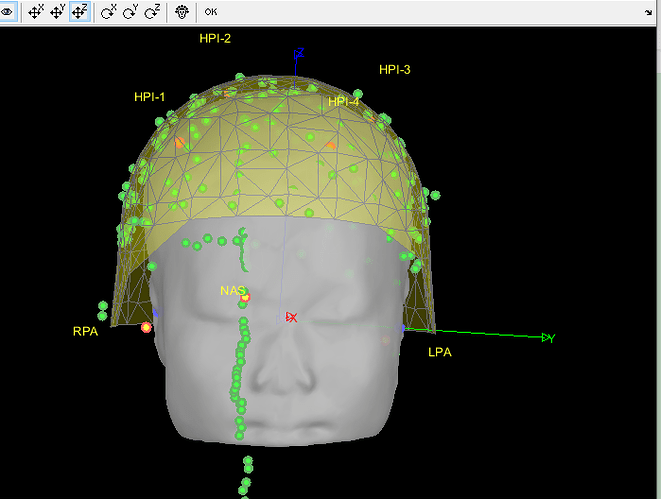
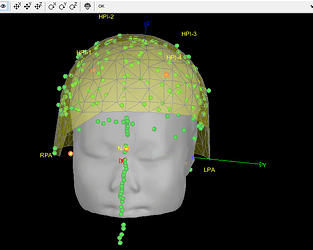
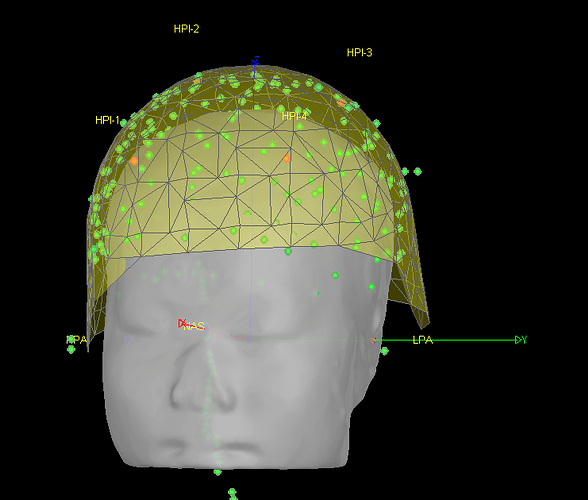
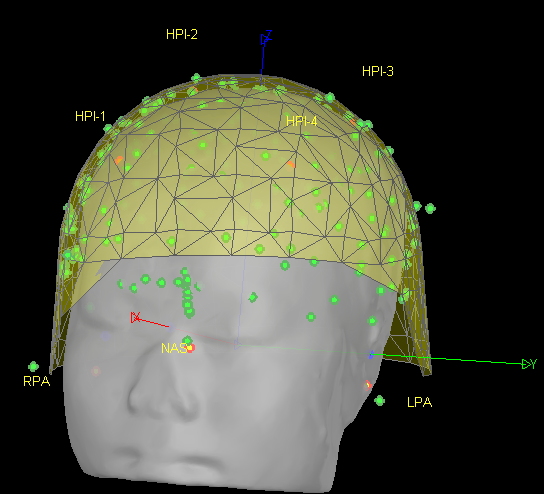
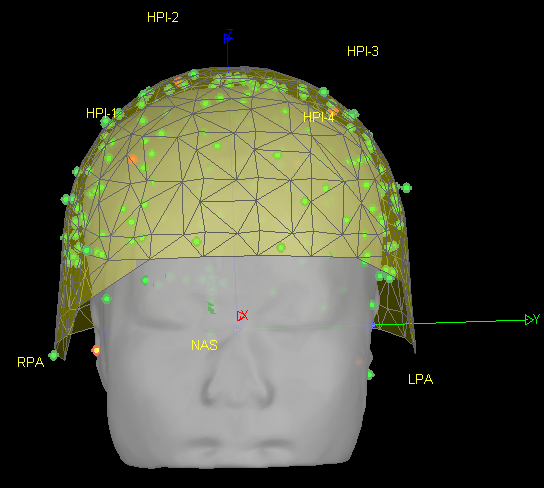
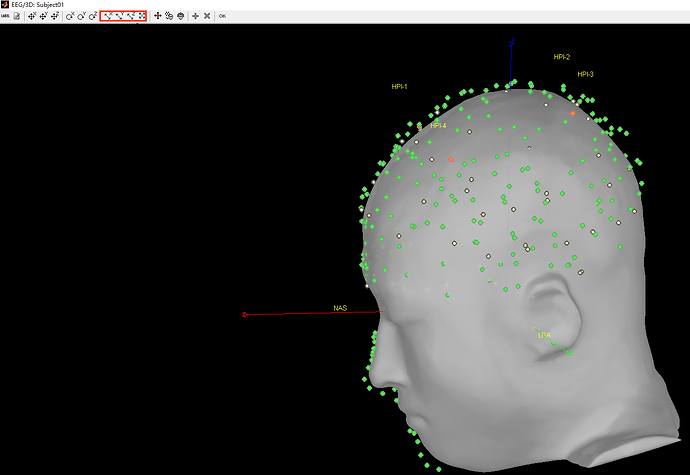
- As can be seen from the figure above, the MEG helmet was tilted to the right . Does this mean that I have to perform manual registration? After the manual registration, the final results are shown in the follwing pictures. It looks like the NAS is on the left. Is that reasonable?
This is up to you to decide, you should know where the subject was in the MEG OR trust your methodology. Unfortunately, we can't magically fix poor acquisition conditions or procedures.
Either you trust the NAS/LPA/RPA fiducials in the FIF files (which are already correctly taken into account without you trying to do any additional stuff with FieldTrip functions), or you trust the head shape (which looks noisy, and probably too large - as I mentioned in my previous post), or you consider that none of this stuff works and you will try to adjust manually the position of the head in the helmet.
Just remember that what to want to see is the real position that the participant had when he/she was sitting in the MEG, not what you think looks better. If the head of the subject was titled, or far from down away from the sensors, or if he/she was moving a lot: there is nothing you can do about that.