Hi,
I would like to ask a couple of questions that two groups at MIT need assistance. Both groups currently do MEG studies without MRI scans.
-
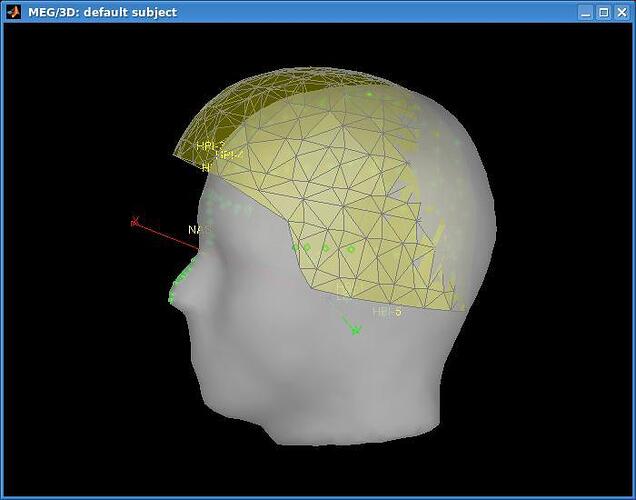
Even though we acquire the 3 fiducial points and extra scalp polhemus points, the data registration with the default anatomy is most of the times very bad. A representative example is this:
http://dl.dropbox.com/u/4202951/defaultsubject_misregistration.jpg
Refining the registration with the polhemus points does not help. Does this happen often with your subjects? I cannot believe that most of our subjects are so different than the default brain, but I do not know what we are doing wrong.
-
Both groups are familiar with EEG data processing and typically average channels across subjects. Even though this is straightforward with EEG channels, it does not make sense to average MEG channels because subjects are positioned differently inside the helmet. Does brainstorm have some way to normalize for this? This would probably involve projecting the data to the brain, shifting the brain to common fiducials and then applying a new forward model to create virtual measurements.
Thank you!
Dimitrios
Hi Dimitrios,
At our site, we routinely digitize the three fiducials, the hpi coils, the 10-20 eeg array, then add another 100 points digitized over the entire upper hemisphere of the head, using the Polhemus Isotrak.
I usually have great success in simply warping these headpoints to the MNI scalp in BrainStorm, then using the corresponding warped cortex as my imaging surface.
How many points are you digitizing?
Are you warping, or trying to align, or both? I generally just go straight to warping in these instances.
– John
Hi John,
Thanks for the reply. I had a look at the data of another researcher and overall they fit better that the ones I was checking earlier. It seems I need to do more effort in training the users how to use the polhemus system.
Regarding the second part of my question, (creating virtual MEG channel measurements from a fixed fiducial location) I thought of way to do it. We could follow the typical forward/inverse modeling to the standard brain for every subject separately, and then multiply each current density map with a predefined fixed forward operator. I will probably need to explain users how to do it here, because I do not believe brainstorm currently supports creating virtual MEG measurements.
Best,
Dimitrios
Regarding the Polhemus, the Neuromag software for the Isotrak includes a “Check” and “Redo” option at the time of acquisition of the fiducials and the HPI Coils.
We have found it essential to do this check, to immediately discover errors. I suggest you make it part of your procedure.
We routinely get “check” results of less than 2 mm; if not, we “redo” until we get it right. Only after both the fiducial set and the hpi set have passed this procedure do we gather the hundred additional scalp points.
Hi Dimitrios,
Sorry for the delay…
-
The picture you sent shows that the subject had a very small head compared with the default Colin27 head. You can see that the green dots look well localized in the helmet, the problem is that the default head is way to big. You need to create a pseudo-anatomy for your subject.
=> Right-click on any channel file for your subject > Head points > Warp > Deform anatomy to fit these points
-
The functions are not that difficult to write. This functionality (co-registration of different MEG runs) is pretty high on my priority list, but not at the top of it. Can you wait for 2-3 months ?
=> If you can’t, you can try to implement it by yourselft as a “process”, I’d be happy to integrate that in Brainstorm =)
Cheers
Francois
Hi Francois!
- The functions are not that difficult to write. This functionality (co-registration of different MEG runs) is pretty high on my priority list, but not at the top of it. Can you wait for 2-3 months ?
=> If you can’t, you can try to implement it by yourselft as a “process”, I’d be happy to integrate that in Brainstorm =)
Is this something that is now available by any chance?
AnneSo
Hey
Yes it is, but not documented yet. We still need to evaluate better the method, but it should work to correct for small movements (< 1 or 2 cm):
Process > Standardize > Co-register MEG runs
In general, all the main new improvements are listed on this “What’s new” page:
http://neuroimage.usc.edu/brainstorm/News
Francois
PS: This is not correcting for continuous head movements yet.
It combines runs with one head position per run.
Thank you François! Do you think we could use it to process several runs over several subjects?
The goal is to produce grand averages at the sensor level over several runs from several subjects. Do you think that this method is appropriate? if not, do you have any other thoughts?
If you want to compute grand averages at the sensor level, it’s probably the best way to go.
If you want averages of sources, it’s probably more accurate to average at the source level.