Hi all,
I am having an issue with aligning an anatomical MRI with my MEG data.
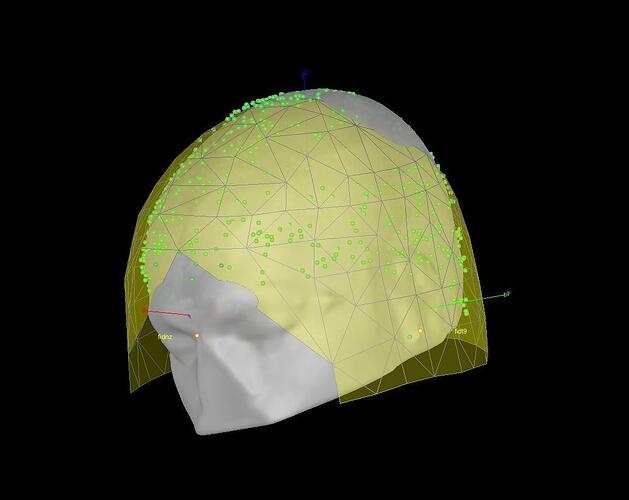
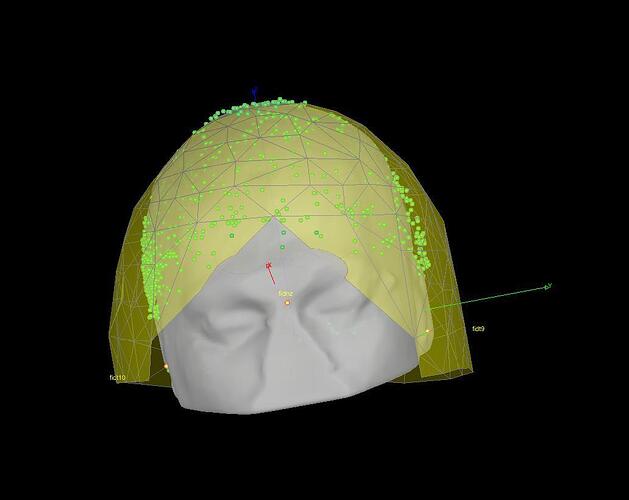
Specifically, it seems to me that the mapping between the digitized head shape and the MEG helmet is wrong (see attached figures).
It looks to me as if the digitized head shape (Polhemus) is inverted compared to the shape of the MEG helmet.
I have checked and double checked the MEG marker positions and the positions of the digitized points, but they seem correct to me.
Any idea what could cause this issue?
I have compared the markers and points of this data set with that of another data set for which the alignment did work. The only notable differences is that
the subjects differ in the extent their heads were tilted backwards (or forwards), i.e. rotated around the y-axis.
I am just speculating, but can it be the case that this produces a change in the orientation calculated by rot3dfit?
This seems to be the function that is used to estimate the transformation matrix R and T to transform the head points to the MEG helmet, but I see no way in which
this function would take into account the position of the scalp relative to the digitized points.
I hope someone can help me out,
My best,
Barrie Klein
Hello,
This is most likely due to an error in the manipulation during the MEG acquisition.
The relative position of the head shape points (green dots) and the MEG sensors (yellow surface) is never altered by Brainstorm. If the two are not aligned, there is probably something that went wrong during the digitization itself (left and right fiducials that were not acquired in the correct order).
If the basic registration, based on the fiducials (NAS, LPA, RPA) looks OK, simply use this one. When you link the recordings to the database, answer NO when asked whether to refine the registration with the head points.
Francois
Dear Francois,
We have checked and double checked the acquisition order of the digitized head points and also the position of the MEG markers. The head points were acquired in the correct order, according to the acquisition software and the MEG markers have relative positions that match their labels. Besides, the MEG scanner should warn us about incorrectly placed markers on the subject’s head. So we are not convinced that this is were the incorrect registration comes from.
After some digging and comparing with another data set that results in a correct alignment, we noticed that the transformation matrices returned by rot3dfit are different for these two data sets:
Transform of correct alignment:
0.8401 -0.10105 -0.53294 -0.020806
-0.051399 -0.9929 0.10723 -0.12066
-0.54 -0.062694 -0.83933 -0.12516
0 0 0 1
Transform of the incorrect alignment:
0.50215 0.026542 -0.86437 -0.067822
-0.070449 -0.99495 -0.071478 -0.10894
0.86191 -0.096786 0.49774 -0.018526
0 0 0 1
Importantly, these matrices have determinants of opposite sign (-1 for the incorrect transform and 1 for the correct transform).
Combined, we believe this indicates that the incorrect transformation matrix has a reflection along one of its dimensions, most likely the z/superior-inferior dimension, compared to the transformation matrix from the other data set.
We think this may be caused by the fact that both the MEG markers and digitized fiducial points lay on a relatively flat 2 dimensional plane. When computing the transformation between these two coordinates, actually two transformations can produce a good alignment between the MEG markers and digitized fiducials of which one is the mirror reflection of the other matrix. We think that this is happening here; the mirrored reflection of what the transformation actually should be does a better job of aligning the MEG makers and digitized fiducials and is thus returned by rot3dfit.
Both data sets were acquired using the same head scanner, MEG scanner, and colected by the same people.
What do you think about this?
My best,
Barrie Klein
Dear Francois,
I had a closer look at this alignment issue yesterday and am more convinced that mirroring/reflecting the digitized head points has to do with this.
I add a point to the head points that is located along the normal in the direction of the head shape, so this point is orthogonal to the plane through the head points. I do the same thing for the MEG marker points, but add a point in both directions, as I do not know where the head is relative to the MEG marker points. So this creates two sets of MEG marker points, one with the added on one side of the plane and one with the added point on the other side of the plane. Then, I recompute the transformation matrix between the 'extended’ head points and both MEG marker point/sets. One these transformation matrices is exactly the same as originally computed by brainstorm (the wrong one that results in the alignement I shared earlier) and the other produces a much more accurate alignment. I can share the code that I have used to do this if you like. Attached is a figure of the resulting alingment.
Apart from this, I also tried swapping the digitized head points, correcting for any incorrect order while acquiring the head points. None of the possible swaps results in an alignment that looked as good as the attached one.
Hope this helps,
My best,
Barrie