Hi team,
I would like to understand if there is a feature equivalent to Fieldtrip's MNI Standard Grid (https://www.fieldtriptoolbox.org/example/sourcemodel_aligned2mni_atlas/). I have individual MRI for all subjects and would like to compare source level information amongst them.
Thanks
Hello,
Indeed there are features for group-level source analysis, warping anatomy to templates, etc. Please have a look at the tutorials, for example https://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects
And one of the steps during anatomy import: https://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#MNI_normalization
Best,
Marc
Thank you so much! Upon reading, I understand the Volume Source model as the closest match to the MNI Standard Grid.
Hello,
I have been experimenting with MRI Volume Head Model. I am using a template grid for all my subjects with 1 cm resolution. Thereby having 1645 grid points.
I performed CAT12 segmentation twice on a single subject using 1900 and 15000 vertices respectively. I computed the MRI Volume Head Volume and generated Inversion model using dSPM.
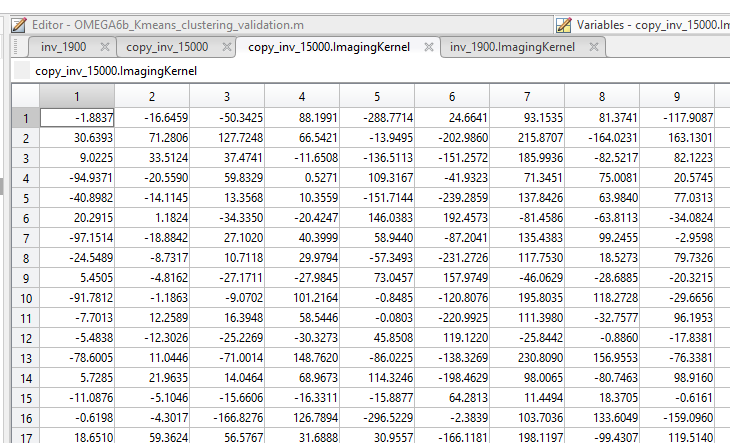
I observed that upon exporting the inverse matrix, the Imaging Kernel has different values for different number of vertices despite having same grid points.
I understand that the vertices are the source dipole and grid points merely average the vertices in their grid. Is that right ?
Also, I wanted to check if there is any way in computing PSD's of the individual grid points rather than scout based.
Thanks
There are 3 perpendicular dipoles per grid point ; hence the head model matrix will comprise 3 times more elements than the number of grid points.
And yes, you can ask Brainstorm to compute the PSD of each grid source
Yes. If each grid point consists of 3 perpendicular source dipoles. Then, I understand that the ImagingKernel should have been same for 15000 vertex CAT12 segmented and 1900 vertex CAT12 segmented inverse head models in the same subject.
However, I see it different in the above results. Can you help me understand of this disparity ?
Can you please refer me the steps? I can't find it in the tutorials. PSD of scout based is explained.
I believe that when you run the PSD process, you have the option of not selecting any scout, which means that it will run the analysis over the entire brain.
I went through the tutorials - vertices are dipoles on the cortex surface. However, I am not able to figure out how vertices combine to form a grid point output ? as seen above.