Hi there,
I have a custom script that could really use the "group files" option of the "process_average.m" script. Is it possible to modify (after copying and pasting to another folder) the "process_average.m" script as a custom process so that when I select "Arithmetic average: mean(x)" it doesn't give me the average but gives me the output from a custom calculation that I wrote while taking into account the group files function?
That is, I only need the "group files" option from the "process_average.m" script and currently looking for a convenient and fast way to implement this to my custom script.
Any suggestions would be most welcome 
Many thanks for your time!
If you only need the grouping, you don't need to modify the function process_average.m file, or copy anything from it. If you are writing your script as a process, you could directly call the function SortFiles from process_average.m. To get an example of how this is called, see line 173 in process_average.m.
[iGroups, GroupComments, GroupNames] = process_average('SortFiles', sInputs, avgtype);
For help with writing scripts in Brainstorm:
https://neuroimage.usc.edu/brainstorm/Tutorials/Scripting
The API for writing processes:
https://neuroimage.usc.edu/brainstorm/Tutorials/TutUserProcess
Thank you.
After the files have been grouped and processed how would you specify accurately where to save the resulting file(s) and register the corresponding file(s) in the database? (i.e. "Everything", "By subject", etc)
Currently, lines 492 - 505 in process_average.m seems like what I need.
> if strcmpi(sInputs(1).FileType, 'data')
> allFiles = {};
> for i = 1:length(sInputs)
> [tmp, allFiles{end+1}, tmp] = bst_fileparts(sInputs(i).FileName);
> end
> fileTag = str_common_path(allFiles, '_');
> else
> fileTag = bst_process('GetFileTag', sInputs(1).FileName);
> end
> OutputFile = bst_process('GetNewFilename', bst_fileparts(sStudy.FileName), [fileTag, '_average']);
> % Save on disk
> bst_save(OutputFile, sMat, 'v6');
> % Register in database
> db_add_data(iStudy, OutputFile, sMat);
What do you think?
The lines of code you identified create a new unique file name for the output file, based on the path of the the folder sStudy.
This folder is identified based on the list of inputs by function GetOutputStudy.
For example, if all the input files are in the same folder, it would return this folder. If all the input files are in the same subject, it would return the @intra folder for this subject. If they are from different subjects, it would use a group analysis subject, etc.
process_average, line 390:
% Get output study
[sStudy, iStudy, Comment, uniqueDataFile] = bst_process('GetOutputStudy', sProcess, sInputs);
If you don't need as many cases to be handled, or if you know exactly where you need to save the output file, maybe you don't need this.
Thank you for the reply.
It looks like the line that you've cited comes before the step of creating a new data file structure (e.g. lines that say sMat.Std) but after processing the data (i.e. averaging the data).
Therefore the following code makes sense to me (I based my code on process_example_customavg.m).
What do you think?
sInputs(iGroups{i}) is due to the below code being within a nested for loop for the SortFiles Function.
% ===== PROCESS =====
% This is where the actual process of data manipulation and calculation takes place.
EpochW = ones(epochSize(1),1,length(sInputs(iGroups{i}))./var((AllMat),0,2)); % Calculation of epoch specific weighting.
AllMat = EpochW.*(AllMat); % Application of epoch specific weighting to all epochs.
AllMat = sum(AllMat,3)./sum(EpochW,3); % Calculation of weighted averaging through the weighted epochs.
% ===== SAVE THE RESULTS =====
[sStudy, iStudy, Comment, uniqueDataFile] = bst_process('GetOutputStudy', sProcess, sInputs(iGroups{i}));
iStudy = sInputs(iGroups{i}).iStudy;
% Create a new data file structure
DataMat = db_template('datamat');
DataMat.F = AllMat;
DataMat.Comment = sprintf('WAvg: 1 (%d)', length(sInputs(iGroups{i}))); % Names the output file as 'WAvg' with the number of epochs used to generate the file.
DataMat.ChannelFlag = ones(epochSize(1), 1); % List of good/bad channels (1=good, -1=bad)
DataMat.Time = Time;
DataMat.DataType = 'recordings';
DataMat.nAvg = length(sInputs(iGroups{i})); % Number of epochs that were averaged to get this file
% Create a default output filename
OutputFiles{1} = bst_process('GetNewFilename', fileparts(sInputs(iGroups{i}).FileName), 'data_WAvg');
% Save on disk
save(OutputFiles{1}, '-struct', 'DataMat');
% Register in database
db_add_data(iStudy, OutputFiles{1}, DataMat);
You are overwriting the iStudy variable returned by GetOutputStudy with the iStudy from the first file in the current group.
It probably had no impact on the code behavior if you don't use iStudy anywhere after that, but I'd recommend you fix this. Maybe by removing the line iStudy = sInputs(iGroups{i}).iStudy?
As long as it works for you, and saves the file in an appropriate folder, you're probably OK.
Thank you, Francois.
After numerous attempts to implement the suggested codes I tried running my process several times but the following error report is what I get.
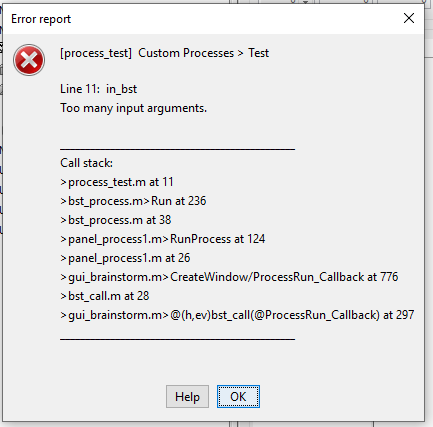
What I find hard to understand is how can there be "too many input arguments" at "Line 11: in_bst"?
Line 11 in question is the following code.
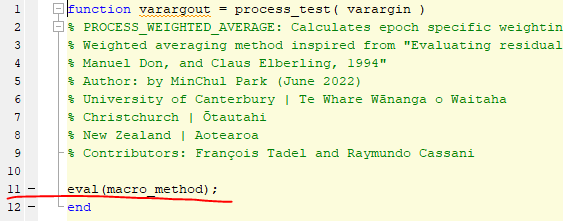
Secondly, how do I understand all the lines below "Call stack"?
Is there some separate Brainstorm tutorial that goes through how to interpret error reports?
I thought implementing "Group files" and process_average, line 390 within my custom code would be a simple task but it's more difficult than I suspected.
Many thanks for your help! 
This call to eval(macro_method) calls the function name passed as the first argument in the previous call in the stack trace. See what is in bst_process at line 236.
It is where the Run function is called: https://neuroimage.usc.edu/brainstorm/Tutorials/TutUserProcess#Sub-functions
Unfortunately, recent versions of Matlab now sometimes fail to trace correctly when there are errors occurring in the eval call.
To dig into what fails in the execution of your process, the best possible tool is the Matlab debugger. Add a breakpoint at the line where you call the function in_bst (= click on the line number), then run the execution of your process. When the execution stops at the requested line you can explore the contents of the variables, navigate in all the call stack, and execute the code line by line using the "Step" and "Step into" buttons.
If the behavior of the debugger is not intuitive to you, start by reading or watching some introduction tutorial to the Matlab debugger. If you're planning to do some more Matlab programming, it will be time well invested.
Is there some separate Brainstorm tutorial that goes through how to interpret error reports?
The error you get here is a Matlab error, unrelated with Brainstorm: you have too many parameters in the call to in_bst. You can google for full error message with double quotes to try to get help from developers forums.
I thought implementing "Group files" and process_average, line 390 within my custom code would be a simple task but it's more difficult than I suspected.
Hopefully you'll develop some new useful skills 
![]()

