Thank you very much for your help, I downloaded the github version of brainstorm in your reply, and then I can now use cat12 to do MRI segmentation.
But if you use fieldtrip to do MRI segmentation, you will still get an error.
The mri data is bug tmp1 IPCAS_age10_brain_template.nii.gz.
BST> FieldTrip install: /home/jade/app/eegpackage/fieldtrip-20201205
the input is volume data with dimensions [182 218 182]
Warning: adding /home/jade/app/eegpackage/fieldtrip-20201205/external/spm12 toolbox to your MATLAB path
using 'OldNorm' normalisation
Warning: adding /home/jade/app/eegpackage/fieldtrip-20201205/external/freesurfer toolbox to your MATLAB
path
Smoothing by 0 & 8mm..
Coarse Affine Registration..
Fine Affine Registration..
performing the segmentation on the specified volume, using the old-style segmentation
SPM12: spm_preproc (v4916) 19:11:07 - 19/12/2020
Completed : 19:11:25 - 19/12/2020
creating scalpmask ... using the anatomy field for segmentation
smoothing anatomy with a 5-voxel FWHM kernel
thresholding anatomy at a relative threshold of 0.100
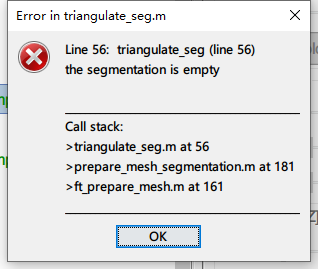
** Error: Line 28: Undefined function 'imfill' for input arguments of type 'double'.
**
** Call stack:
** >volumefillholes.m at 28
** >ft_volumesegment.m at 619
** >process_ft_volumesegment.m>Compute at 241
** >process_ft_volumesegment.m>ComputeInteractive at 389
** >process_ft_volumesegment.m at 27
** >bst_call.m at 28
** >tree_callbacks.m>@(h,ev)bst_call(@process_ft_volumesegment,'ComputeInteractive',iSubject,iAnatomy) at 1059
**
In addition, if I use brainstorm to generate BEM, an error will be reported.
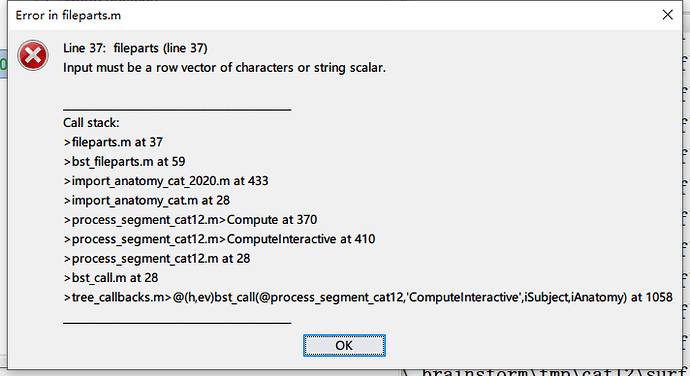
** Error: Line 185: scatteredInterpolant
** The input points must be a double array.
**
** Call stack:
** >griddata.m>useScatteredInterp at 185
** >griddata.m at 126
** >tess_parametrize_new.m at 48
** >tess_bem.m at 133
** >process_generate_bem.m>ComputeInteractive at 152
** >process_generate_bem.m at 24
** >bst_call.m at 28
** >tree_callbacks.m>@(h,ev)bst_call(@process_generate_bem,'ComputeInteractive',iSubject,iAnatomy) at 1050
**
Then if you use fieldtrip to generate BEM, an error will be reported. And this error will also report the same error when I use John E. Richards' mri template to generate bem. (I don’t know if I need to send you a John E. Richards mri template example?)
BST> FieldTrip install: /home/jade/app/eegpackage/fieldtrip-20201205
the input is volume data with dimensions [182 218 182]
using 'OldNorm' normalisation
Smoothing by 0 & 8mm..
Coarse Affine Registration..
Fine Affine Registration..
performing the segmentation on the specified volume, using the old-style segmentation
SPM12: spm_preproc (v4916) 19:19:39 - 19/12/2020
Completed : 19:19:55 - 19/12/2020
creating scalpmask ... using the anatomy field for segmentation
smoothing anatomy with a 5-voxel FWHM kernel
thresholding anatomy at a relative threshold of 0.100
** Error: Line 28: Undefined function 'imfill' for input arguments of type 'double'.
**
** Call stack:
** >volumefillholes.m at 28
** >ft_volumesegment.m at 619
** >process_ft_volumesegment.m>Compute at 241
** >process_ft_volumesegment.m>ComputeInteractive at 389
** >process_ft_volumesegment.m at 27
** >process_generate_bem.m>ComputeInteractive at 154
** >process_generate_bem.m at 24
** >bst_call.m at 28
** >tree_callbacks.m>@(h,ev)bst_call(@process_generate_bem,'ComputeInteractive',iSubject,iAnatomy) at 1050
**
Thank you very much for your help and look forward to your reply.