Dear all,
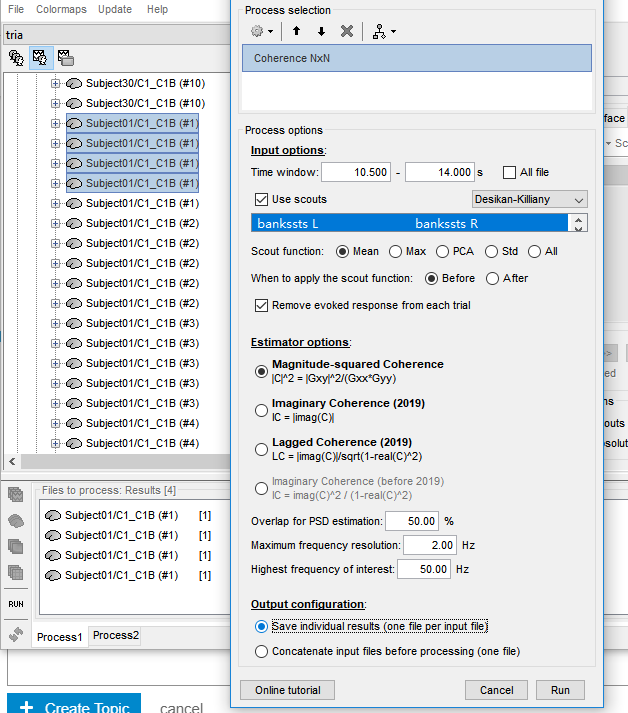
I first computed the coherence on single trials at the source level using Mininum norm imaging dSPM within the DK scout atlas.
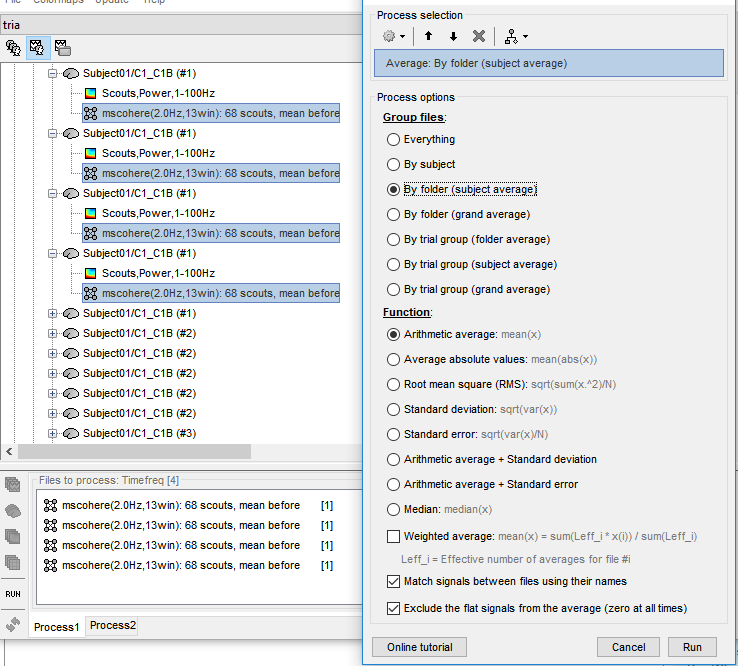
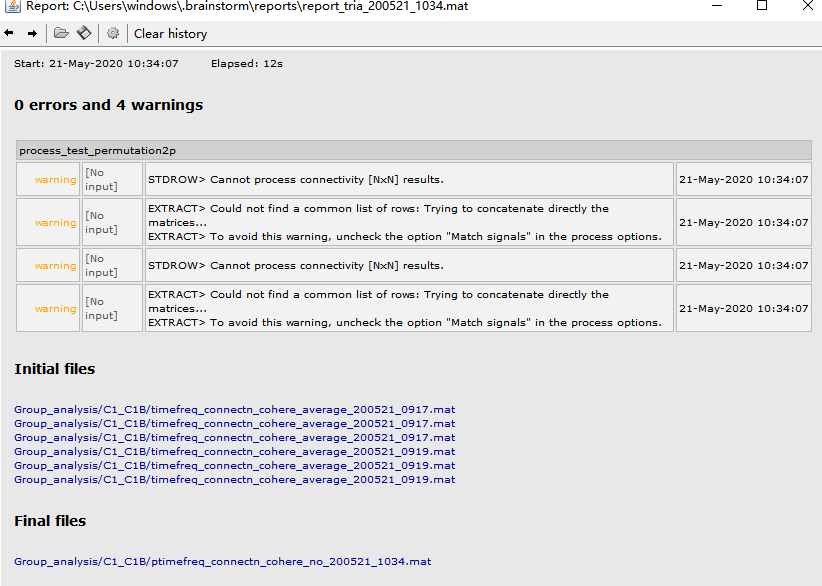
Then, I averaged the coherence files of each subject under each condition. An warning appeared.
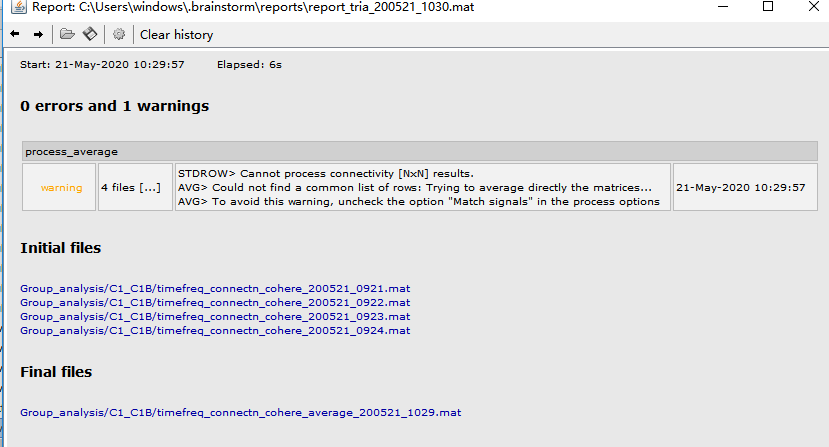
I didn't mind the warning and conducted paired permutation T test, warning appeared again 'Cannot process connectivity [N*N] results', 'Could not find a common list of rows'...
what's wrong with my process?
Uncheck the "Match signals between files using their names" option

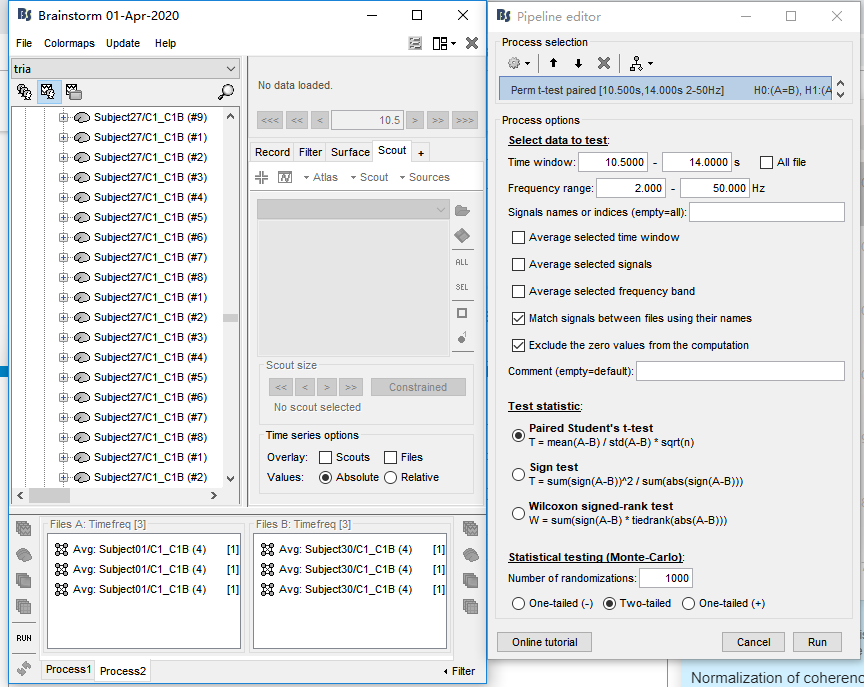
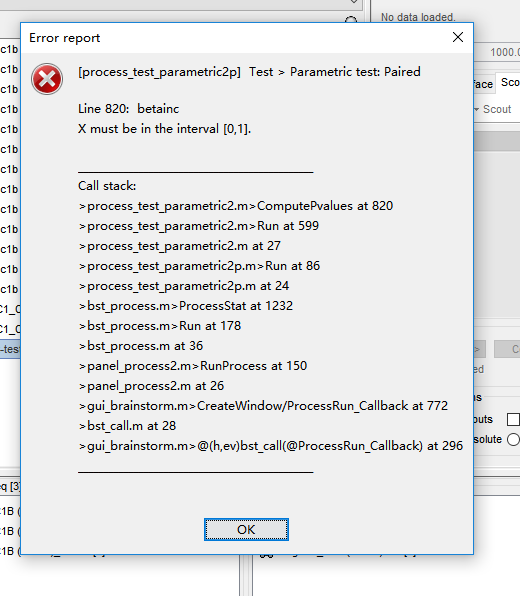
Many thanks, I unchecked the 'match signals between files using their names' option. It works well for averaging and permutation t test. However, an error occurred when I conducted paired t test.
Is there anything wrong?
Update Brainstorm and you'll get access to a new option "Output configuration: Save average connectivity matrix (one file)" directly in the coherence process options.
However, an error occurred when I conducted paired t test.
This error means that most likely there is no meaningful data in input, but it should be caught earlier and Brainstorm should display a nicer error message than this... Do you also get this when you try with your real data (ie. the real data you want to test)?
Please note that you should probably not be using parametric tests for coherence... For data data is not normally distributed, prefer non-parametric tests:
https://neuroimage.usc.edu/brainstorm/Tutorials/Statistics#Nonparametric_permutation_tests
Many thanks Francois, I tried with the real data I want to test, no error occurred this time. Sorry for another question: When I computed the N*N coherence at the source level, should the source be rectified before the coherence computation?
When I computed the N*N coherence at the source level, should the source be rectified before the coherence computation?
No. Applying an absolute value alters the frequency contents of the file and makes coherence analysis impossible.
Note that connectivity analysis at the source level might be very complicated, and does not necessarily give more information than connectivity analysis and the sensor level. All the information is already present in the recordings, regularized minimum norm solutions destroy some information and introduces a massive amount of linear mixing between the signals, making the connectivity analysis more difficult (more signal, more linear mixing).
Hi Francois,
not necessarily a Brainstorm question, but do you have any papers in mind that you could share with regards to advising against doing connectivity analysis at the source level?
I've often seen the Gross et al. 2013 ("Good Practice for Conducting and Reporting MEG Research") paper cited as why one would want to do connectivity at the source rather than the sensor level, along with a few other papers.
Thank you!
Advise against source-level connectivity analysis?
I don't, but PUBMED knows better