Hi @edelaire @Francois, and team!
I am wondering if you can help me in my analyses.
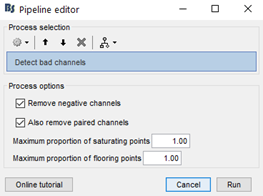
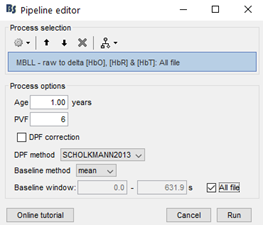
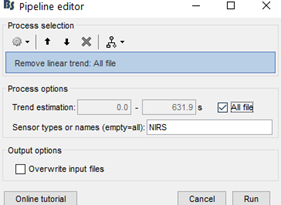
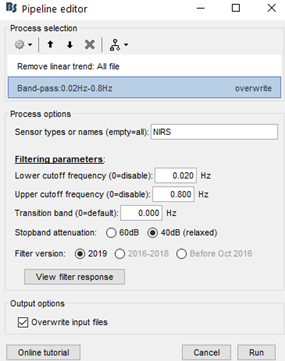
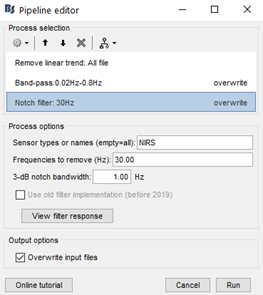
My team and I made fNIS in 61 babies, in four auditory stimuli. All data are already in NIRSTORM. We processed every subjects/conditions with motion correction (manually selected) - detect bad channels - remove glitches - remove linear trend - band pass filter - notch filter - mark flat channels as bad - code in Matlab to detect bad channels due to S/N ratio - visual analyses to detect bad channels and bad trials - standardize the good trials (baseline normalization) and arithmetic average of the trials.
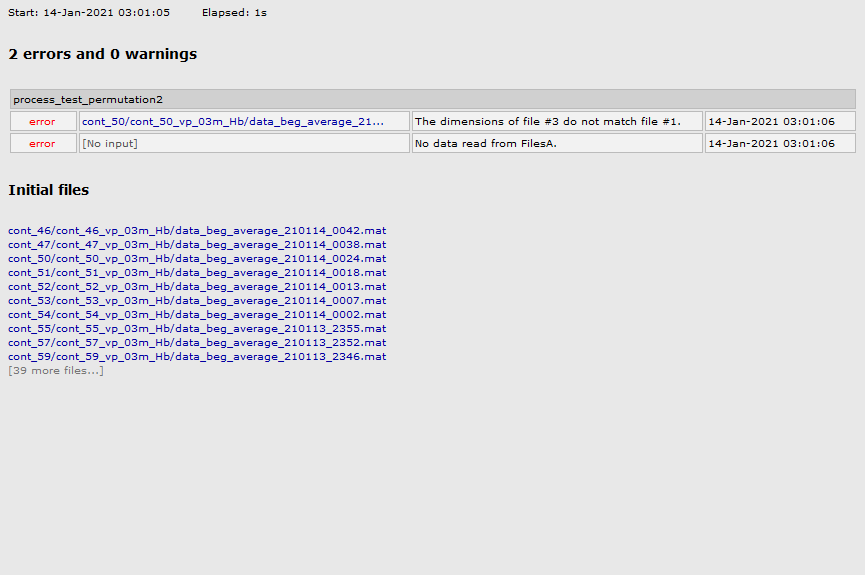
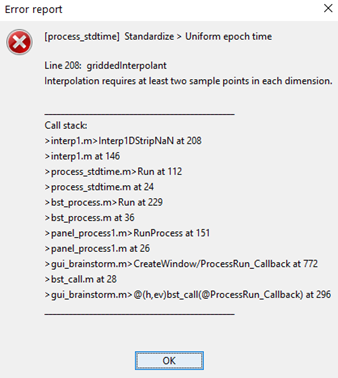
Now I need to make the stats to compare groups (typical and atypical babies) in each condition. I'm trying to run permutation test but I got this error message:
Indeed, I had some "NIRS-BRS sensors" files with 252 and others with 258 (I dont know why, since the geometry file is the same one). I manage to fix it and now I have all files with 252 sensors, but I still get the same error message to run stats.
I already tried “Standardize -> Uniform list of channels”, but “All the input files have identical channel names”. I also tried to delete the files indicated in the error message, or even delete some subjects randomly, but it did not work.
Do you know what should I do to move forward?
Any help would be greatly appreciated!
Kind regards,
Gabriela