Hello everyone, hello Edouard !
I recently came back to NIRSTORM to analyze some data and i had issues with plots and data averaging.
I'm using Brite MKII and MKIII, files are .nirs.
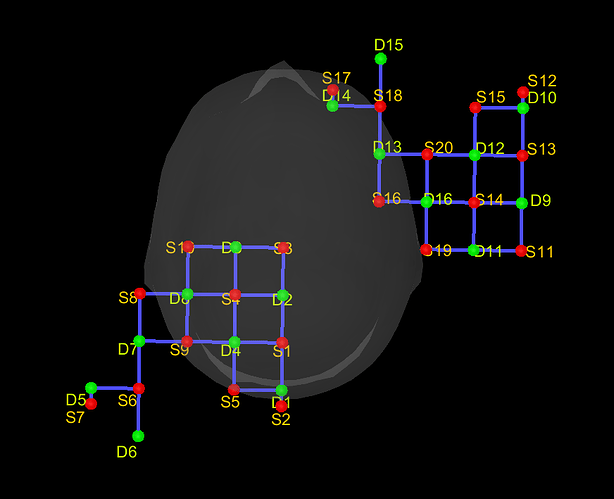
When i import a nirs raw file, my channels are loaded and look like this ( global channel file for all subjects, NIRS-BRS channels (118) ):
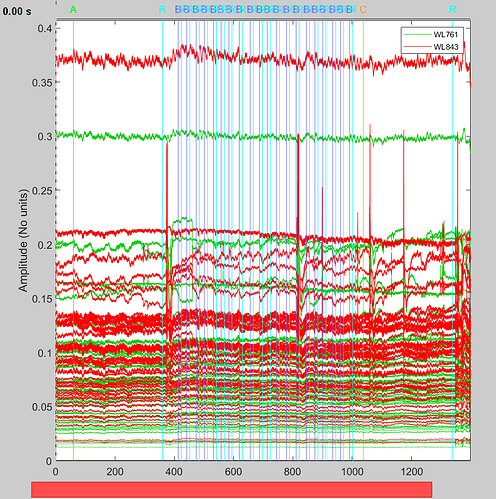
Here i can plot the nirs raw file without any problem :
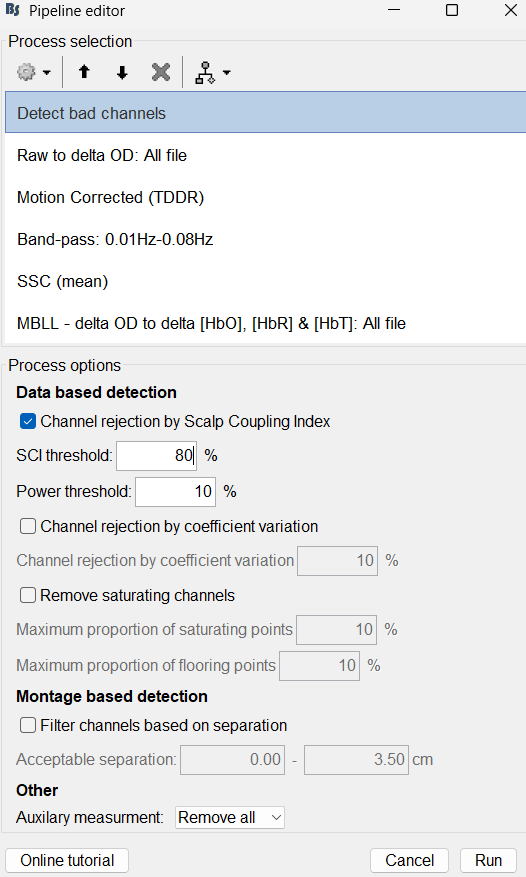
Next i will process the file with the following steps :
Note that channels can be removed by SCI/PSP (i will have another question on this part later ![]() ) and that i removed all NIRS AUX.
) and that i removed all NIRS AUX.
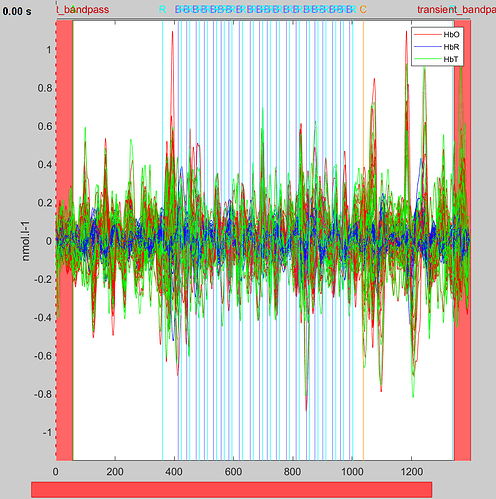
Then, i can plot the resulting concentrations.
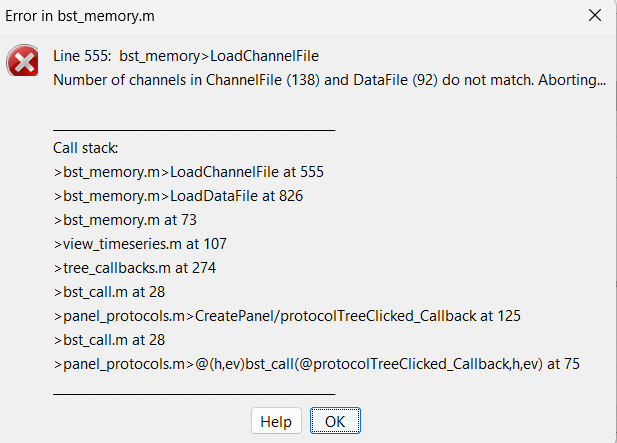
Now, if i want to check each plot in between, after motion correction or bandpass filter, i get the error:
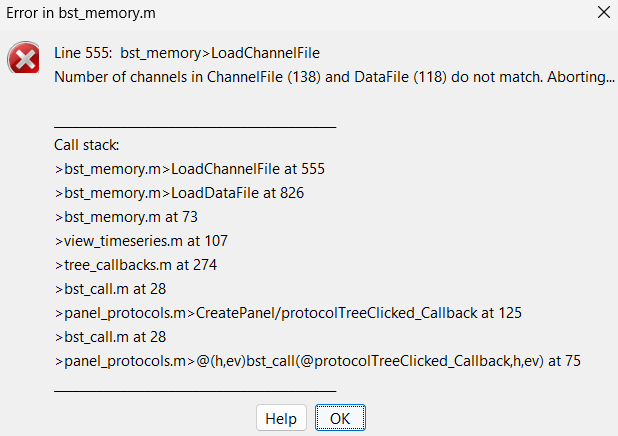
If i understand it correctly, it is because each data corresponds to a channel file but it has been modified by removing channels earlier and after row to conc because it added channels that correspond to HbO / HbR / HbT.
If i try to re open the raw file, i also get the error but it mentions the original number of channels:
So, that is the part corresponding to the plots problem.
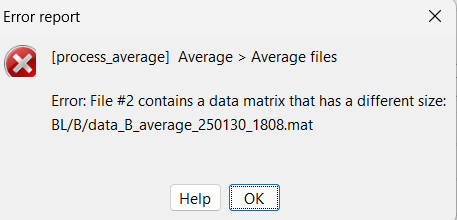
Now, this error also prevents me from averaging files from different subjects.
Because some subjects has "bad channels", they are removed from the channel file linked to their data (again, if i understood it correctly), so when i try to average files from different subjects (corresponding to block average files), it tells me that files have not the same number of channels.
But, if i try this with subjects that had not removed channels, it works perfectly !
For the plots, i can deal with it by re importing the file and do the processing again if i want to check the plot (even if it is not practical), but for the averaging, i can not anything, so it blocks me.
I hope that i made myself understandable and that i provided enough details ! Do not hesitate to tell me if you need more infos etc ![]()
See you, regards,
Brieuc