Hello,
I have successfully completed the 3D reconstruction of my fNIRS signal along the cortical surface by following the tutorials and paper provided by the developers. I am truly grateful to the development and support team for their assistance.
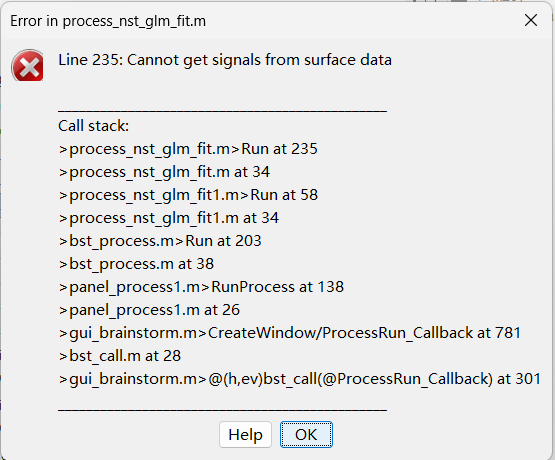
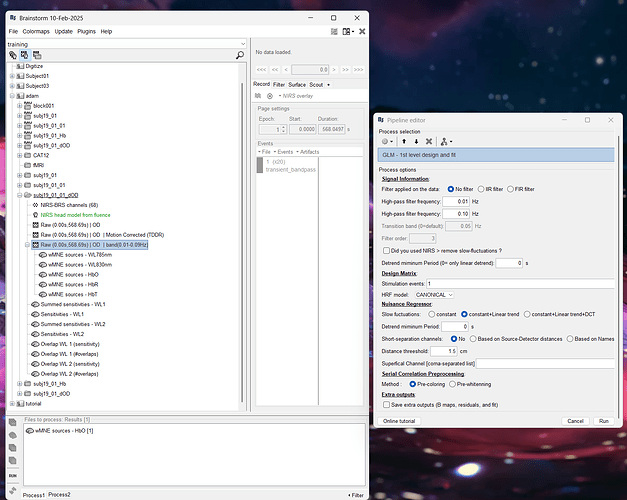
In the paper (https://www.biorxiv.org/content/10.1101/2024.09.05.611463v1), the developers mentioned that the GLM module in NIRSTORM can be utilized after 3D reconstruction along the cortical surface. I attempted to run the GLM based on the "wMNE sources-HbO" to generate the t-value map, but encountered an error stating "can not get signal from surface data". I have attached screenshots of the GLM setup and the error message.
As I have been unable to find tutorials addressing this issue, I would greatly appreciate any assistance in resolving it.
Thank you!!