Dear all,
after using Brainstorm for many year with MEG and EEG data, I would like to explore its capabilities in processing fNIRS data. However, I already have problems importing my raw data. ![]()
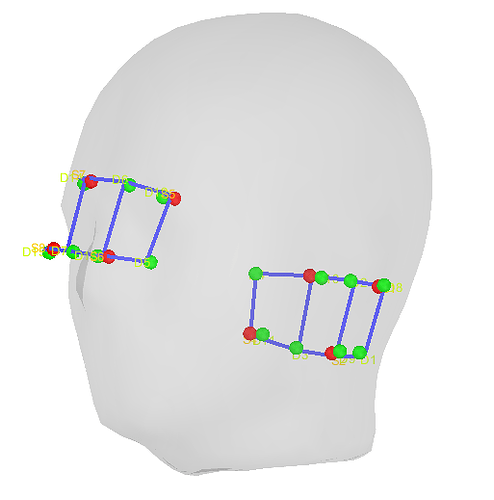
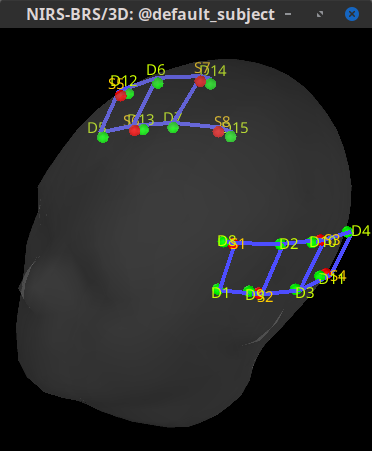
I have a pilot dataset recorded with the NIRx NIRSport 2 system and the Aurora fNIRS 2023.9 acquisition software. When importing the .snirf file into brainstorm, the channel locations are rotated and I do not understand how to correct this. The coordinates given in the channel file correspond with the information provided in the digpts.txt file of the Aurora output. However, I suspect that the reference points may not be compatible.
[Channels should be located over left and right temporal lobes]
Can somebody help me with this? I saw some old discussions on the topic, but those did not really help me. I attached a .snirf file and the corresponding digpts.txt file.
2024-05-28_002.snirf (1.6 MB)
digpts.txt (808 Bytes)
Thanks a lot!
Sebastian